Extended Data Fig. 1. Experimental flow outline and demographic/clinical information for subjects for monocytes isolation.
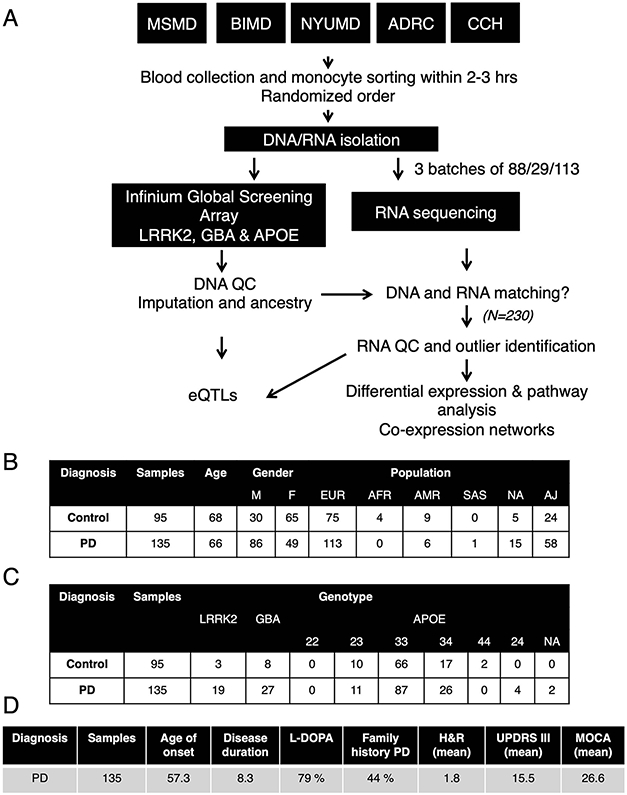
(A) Blood was collected from five independent clinics across New York City (ADRC, CCH, MSMD, BIMD, and NYUMD; details described in Methods) and transferred to the Icahn School of Medicine at Mount Sinai for monocyte sorting and RNA/DNA isolation. Samples were genotyped for common SNPs using Global Screening Array (GSA) and LRRK2, GBA and APOE were independently genotyped. RNA-seq was performed at Genewiz Inc. in three independent and randomized batches. DNA and RNA data was subjected to stringent QC, DNA data was imputed and ancestry was calculated. DNA and RNA were compared to the identification of miss-matches prior outlier identification. After QC, a total of 230 samples were used for subsequent analysis. (B) Demographic, (C) genotype and (D) clinical variables describing the 230 samples included in the study.
