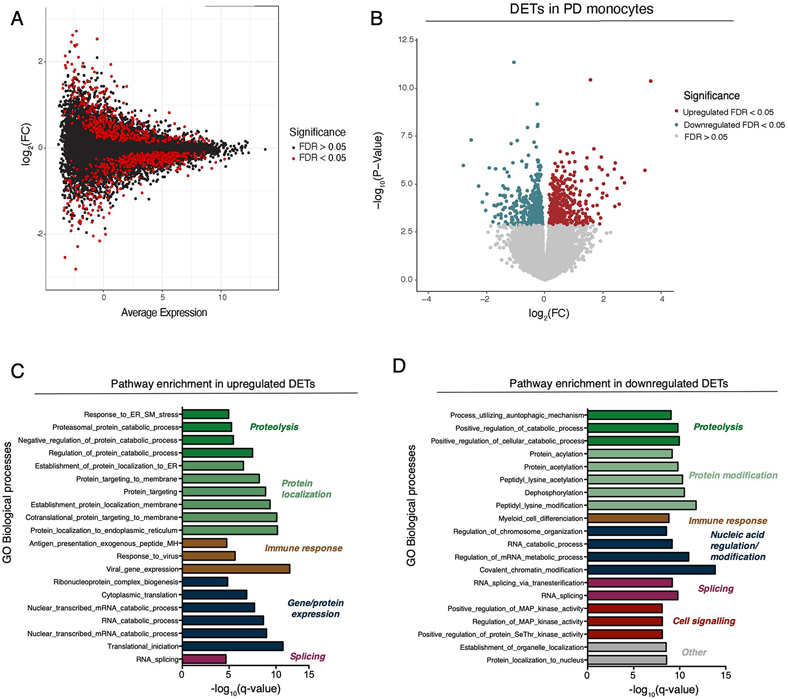
Extended Data Fig. 2. Differential expression analysis at the transcript level in PD and controls derived monocytes.
(A) MA plot showing the fold-change (log2 scale) at the transcript level in the y-axis and the mean of log2counts (x-axis), highlighting the DETs at FDR < 0.05 in red. (B) Volcano plot showing the fold-change (log2 scale) of transcripts between PD-monocytes (n = 135) and controls (n = 95) (x-axis) and their significance in the y-axis −log10 P-value scale). DETs at FDR < 0.05 are highlighted in red (upregulated) and blue (downregulated). Moderated t-statistic (two sided) is used for statistical test (see R package limma). (C) Pathway enrichment analysis for the upregulated (n=230 independent samples) and (D) downregulated DETs using Biological processes from GSEA. Significance is represented in the x-axis (−log10 P-value scale of the q-value). Only the 20 most significant pathways (q-value < 0.05) with a minimum overlap of 5 genes are shown. Pathways are grouped and colored by biological related processes. n=230 independent samples