SUMMARY
Piwi-interacting RNAs (piRNAs) regulate many biological processes through mechanisms that are not fully understood. In Caenorhabditis elegans, piRNAs intersect the endogenous RNA interference (RNAi) pathway, involving a distinct class of small RNAs called 22G-RNAs, to regulate gene expression in the germline. In the absence of piRNAs, 22G-RNA production from many genes is reduced, pointing to a role for piRNAs in facilitating endogenous RNAi. Here, however, we show that many genes gain, rather than lose, 22G-RNAs in the absence of piRNAs, which is in some instances coincident with RNA silencing. Aberrant 22G-RNA production is somewhat stochastic but once established can occur within a population for at least 50 generations. Thus, piRNAs both promote and suppress 22G-RNA production and gene silencing. rRNAs and histones are hypersusceptible to aberrant silencing, but we do not find evidence that their misexpression is the primary cause of the transgenerational sterility observed in piRNA-defective mutants.
Graphical Abstract
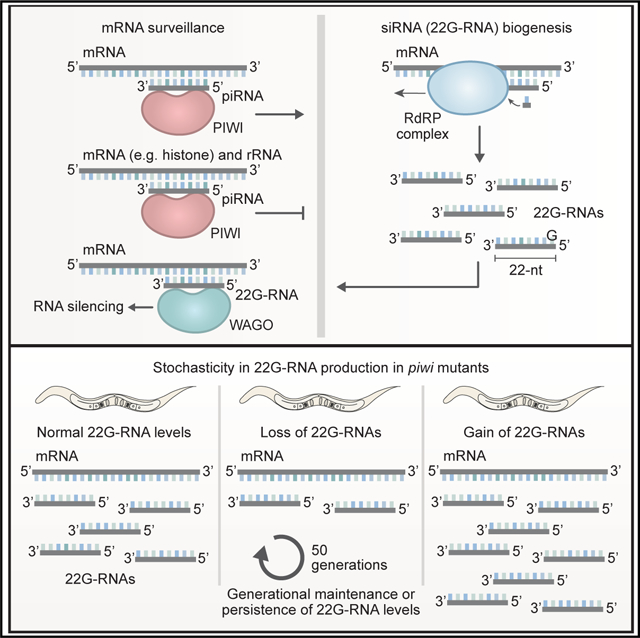
In brief
Montgomery et al. show that piRNAs both promote and suppress siRNA production and RNA silencing in C. elegans. Gain or loss of siRNAs occurs somewhat stochastically in piRNA-defective mutants but once established, it occurs across numerous generations.
INTRODUCTION
Piwi-interacting RNAs (piRNAs) affect many different processes in the germline (Iwasaki et al., 2015). Perturbing the piRNA pathway leads to sterility in many species, in part because of its critical role in silencing transposable elements (Iwasaki et al., 2015). In the nematode Caenorhabditis elegans, loss of piRNAs does not lead to immediate sterility, but instead causes a gradual loss of fertility over numerous generations such that the germline loses its immortal nature (Simon et al., 2014). As sterility ensues in piRNA mutants, the germ granules involved in RNA surveillance collapse (Spichal et al., 2021). However, the sterility of piRNA mutants is not clearly linked to transposon activation and genomic instability (Barucci et al., 2020; Reed et al., 2020; Simon et al., 2014; Spichal et al., 2021). Several models have been proposed to explain the progressive sterility of piRNA mutants. For example, we and others showed that piRNAs prevent silencing of essential genes by an endogenous RNA interference (RNAi) pathway involving small interfering RNAs (siRNAs) (de Albuquerque et al., 2015; Phillips et al., 2015). Recently, the germline mortality of piRNA mutants was attributed to the silencing of essential histone genes (Barucci et al., 2020). In a conflicting report published while this manuscript was under review, sterility was instead linked to aberrant silencing of ribosomal RNAs (rRNAs) (Wahba et al., 2021).
In C. elegans, piRNAs associate with a single Piwi protein, PRG-1, where they act as sequence-specific guides to direct the complex to target mRNAs within germ granules (Batista et al., 2008; Das et al., 2008; Ruby et al., 2006; Wang and Reinke, 2008). The interaction between piRNAs and target mRNAs leads to the production and subsequent amplification of siRNAs, called 22G-RNAs, by an RNA-dependent RNA polymerase in association with a collection of proteins called the Mutator complex (Bagijn et al., 2012; Lee et al., 2012; Phillips et al., 2012). The Mutator complex is seeded by MUT-16 adjacent to the germ granules that house the piRNA machinery (Batista et al., 2008; Phillips et al., 2012). This is the same pathway that is activated by canonical siRNAs produced from double-stranded RNA during exogenous RNAi and is therefore commonly referred to as endogenous RNAi (Almeida et al., 2019a; Claycomb, 2014). piRNA-dependent 22G-RNAs bind a worm-specific class of Argonautes called WAGOs (Gu et al., 2009). A genetically distinct class of 22G-RNAs binds to the Argonaute CSR-1 and is thought to act in opposition to piRNAs to promote or fine-tune gene expression (Campbell and Updike, 2015; Cecere et al., 2014; Claycomb et al., 2009; Conine et al., 2013; Gerson-Gurwitz et al., 2016; Seth et al., 2013; Wedeles et al., 2013).
C. elegans contains thousands of distinct piRNAs that require only partial sequence complementarity to bind target mRNAs (Bagijn et al., 2012; Shen et al., 2018; Zhang et al., 2018). Consequently, piRNAs are thought to engage most, if not all, mRNAs in the germline (Bagijn et al., 2012; Lee et al., 2012; Shen et al., 2018). Furthermore, the 22G-RNAs produced from piRNA targets are thought to serve as a memory of piRNA activity that in some instances can persist in the absence of the initial piRNA trigger (Ashe et al., 2012; Luteijn et al., 2012; Shirayama et al., 2012).
Loss of piwi/prg-1, which results in nearly complete loss of piRNAs, leads to a reduction in WAGO-class 22G-RNA levels from many mRNAs in the germline; however, histones produce elevated levels of 22G-RNAs in prg-1 mutants (Barucci et al., 2020; Reed et al., 2020). Through a genome-wide analysis of 22G-RNAs, we discover that many protein coding genes, as well as ribosomal RNAs (rRNAs), gain 22G-RNAs in prg-1 mutants. Gain of 22G-RNAs in prg-1 mutants was often coincident with silencing of the mRNA from which the 22G-RNAs were derived. Hyperaccumulation of 22G-RNAs was detectable across 50 generations. We show that aberrant production of 22G-RNAs occurs somewhat stochastically, although certain genes, including histones and rRNAs are hypersusceptible to this phenomenon. More generally, both gain and loss of 22G-RNAs occurs inconsistently within populations of prg-1 mutants. Our results indicate that piRNAs have broad roles in both promoting and suppressing 22G-RNA production and gene silencing.
RESULTS
Aberrant 22G-RNA production and gene silencing in prg-1 mutants
We and others recently showed that in C. elegans piwi/prg-1 mutants, histones are directed into an endogenous RNAi pathway where they spawn high levels of 22G-RNAs, which is in some instances coincident with reduced histone mRNA levels (Barucci et al., 2020; Reed et al., 2020). To further explore this phenomenon, we examined 22G-RNA production from all annotated protein coding genes using RNA sequencing (RNA-seq) datasets from dissected distal gonads from wild-type and prg-1 and mut-16 mutants (Reed et al., 2020). Roughly 75% of protein coding genes yielded 22G-RNA reads (at least 1 read) in wild-type animals, and to a somewhat lesser extent in prg-1 and mut-16 mutants, indicative of the prevalence of 22G-RNAs in the germline (Figure 1A; Table S1). A subset of genes produced highly elevated levels of 22G-RNAs in prg-1 mutants (Figure 1B; Table S1). We did not observe elevated levels of 22G-RNAs in animals containing a mutation in henn-1, which also acts within the piRNA pathway but has only a modest impact on 22G-RNA production in the germline (Figure 1C; Table S1) (Billi et al., 2012; Kamminga et al., 2012; Montgomery et al., 2012; Pastore et al., 2021; Svendsen et al., 2019). Nor did we observe elevated levels of 22G-RNAs in mut-16 mutants, in which WAGO-class 22G-RNAs are nearly completely lost, including those that are piRNA-dependent, but which retain CSR-1-class 22G-RNAs (Figures 1A and S1A; Table S1). We therefore do not believe that this phenomenon is related to loss of piRNA-dependent 22G-RNAs or to noise or variation in 22G-RNA production between different strains.
Figure 1. Hyperaccumulation of 22G-RNAs and aberrant gene silencing in prg-1 mutants.
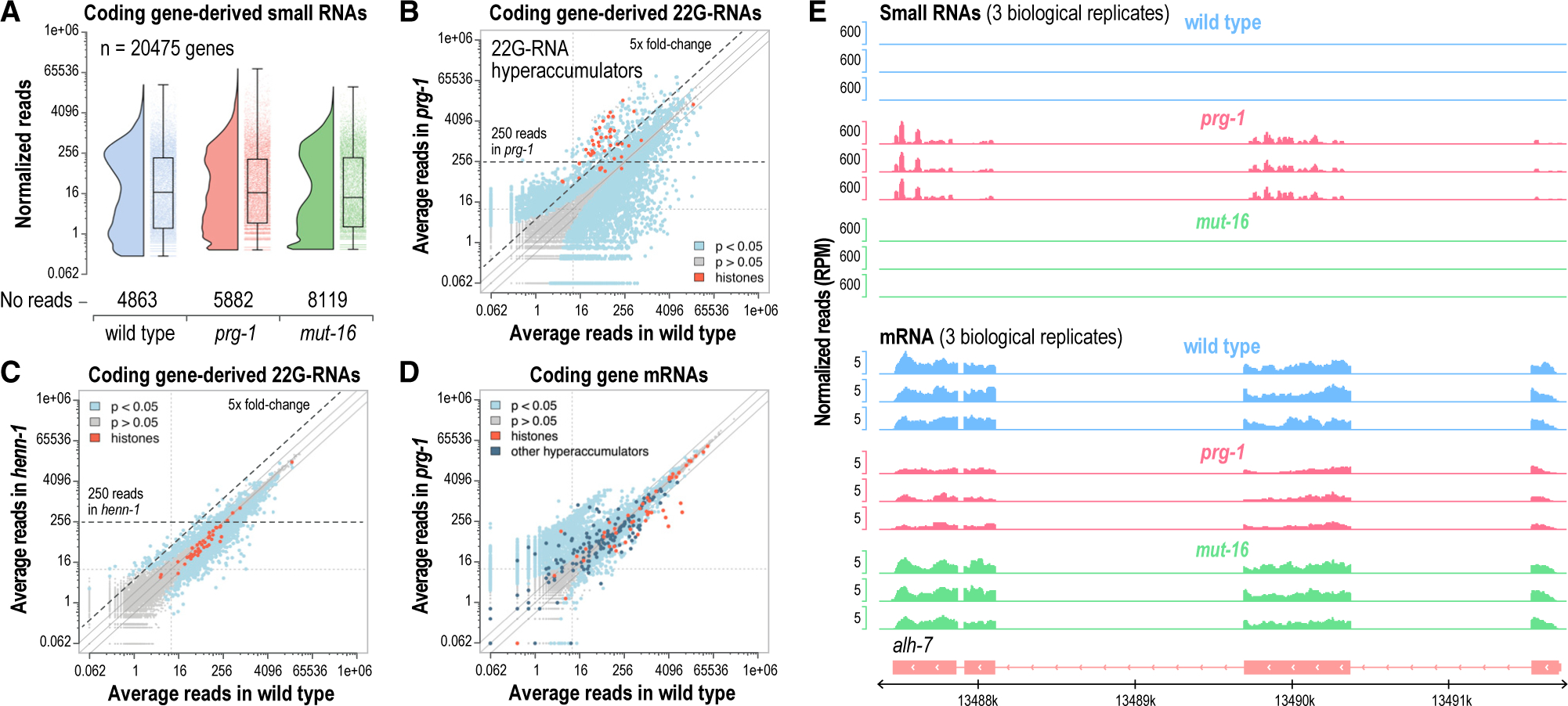
(A) Rain shadow boxplots displaying normalized log2-transformed small RNA high-throughput sequencing reads for each annotated coding gene in dissected distal gonads of wild-type, prg-1(n4357), and mut-16(pk710) mutants (n = 3 biological replicates for each strain). The values on the y axis are reverse transformed to reflect non-transformed values.
(B and C) Scatterplots displaying each gene as a function of normalized log2-transformed small RNA reads in wild-type and prg-1(n4357) (B) or henn-1(pk2295) (C) mutants. The axes are reverse transformed to reflect non-transformed values. Hyperaccumulators are classified as genes that yielded an average of >250 normalized reads and which were upregulated >5-fold in prg-1 mutants, as indicated by the dashed lines. Libraries are from dissected distal gonads (n = 3). Solid lines above and below the y = x lines indicate 2 and −2 fold-changes.
(D) Scatterplot displaying each gene as a function of normalized log2-transformed mRNA high-throughput sequencing reads in wild-type and prg-1(n4357) (axes show non-transformed values).
(E) mRNA and small RNA read distribution across a representative gene locus, alh-7, that hyperaccumulated 22G-RNAs in prg-1 mutants. Three biological replicates are shown for wild-type, prg-1(n4357), and mut-16(pk710).
We identified 222 genes that yielded >250 22G-RNA reads on average in prg-1 mutants and that were upregulated >5-fold relative to wild-type. These arbitrarily applied cutoffs captured most histones and minimized what may be noise in 22G-RNA expression (Figure 1B; Table S1). Of the 222 genes, ~30% were detectably downregulated > 1.3 fold at the mRNA level (Figure 1D; Table S2). However, many genes that gained 22G-RNAs were not misregulated and some were instead upregulated in prg-1 mutants, which may in part reflect the inconsistent relationship between 22G-RNA expression and gene silencing (Reed et al., 2020) (Figure 1D). Twenty-seven of the 66 genes that produced elevated levels of 22G-RNAs and had reduced mRNA levels are histones, which comprised several of the most highly downregulated genes (Figures 1D and S1B; Table S2). The remaining 39 genes are unrelated to histones, such as the aldehyde dehydrogenase alh-7 (Figures 1E and S1B; Table S2). These results are consistent with Barucci et al. (2020), which also identified a prevalence for histones among downregulated genes with elevated 22G-RNA levels in prg-1 mutants. Thus, piRNAs have roles in both promoting 22G-RNA production from some transcripts and in preventing aberrant 22G-RNA production from others.
Hyperaccumulation of 22G-RNAs from rRNAs in prg-1 mutants
We next examined small RNAs produced from long non-coding RNAs and structural RNAs, including small nuclear RNAs (snRNAs), Y RNAs, small nucleolar RNA (snoRNAs), tRNAs, and rRNAs, to determine if they also lose or gain 22G-RNAs in the absence of piRNAs. Many non-coding RNAs produced relatively high levels of 22G-RNAs, a subset of which were depleted in prg-1, as well as in mut-16 mutants, indicating that like many coding genes they are targeted by piRNAs and routed into the RNAi pathway (Figures 2A and 2B; Table S3). However, rRNAs were distinct in having high levels of small RNAs that were elevated >5-fold in prg-1 mutants but which were not substantially affected in mut-16 mutants (Figures 2A and 2B). These small RNAs bare the hallmarks of 22G-RNAs: antisense orientation, preference for a 5’G, and length of 22-nts, and are therefore likely to be the ribosomal siRNAs (risiRNAs) previously associated with rRNA misprocessing, although we do not have evidence that rRNAs are more prone to misprocessing in prg-1 mutants (Figures 2C and 2D) (Zhou et al., 2017; Zhu et al., 2018).
Figure 2. Aberrant production of 22G-RNAs from rRNAs in prg-1 mutants.
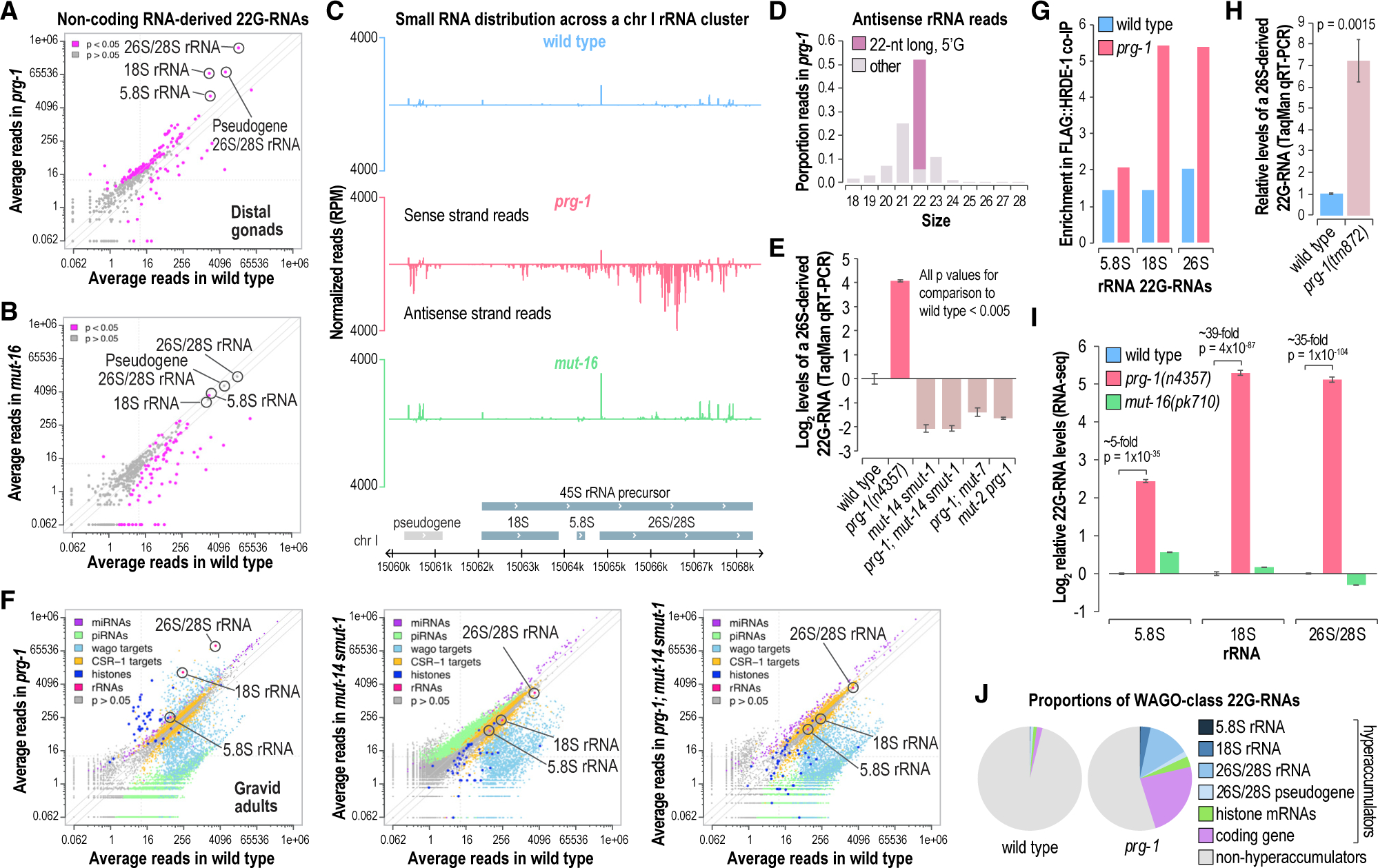
(A and B) Scatterplots displaying each non-coding RNA as a function of normalized log2-transformed small RNA high-throughput sequencing reads in wild-type and prg-1(n4357) (A) or mut-16(pk710) (B) mutants (axes values are reverse transformed to reflect non-transformed values). rRNAs are circled. Libraries are from dissected distal gonads (n = 3 biological replicates for each strain). Solid lines above and below the y = x lines indicate 2 and −2 fold-changes.
(C) Small RNA read distribution across an rRNA locus. One representative of 3 biological replicates is shown for wild-type, prg-1(n4357), and mut-16(pk710).
(D) Antisense rRNA-derived small RNA size distribution in prg-1(n4357) mutants.
(E) Relative log2-transformed levels of a 26S/28S rRNA-derived 22G-RNAs in wild-type animals and various mutants as determined by TaqMan qRT-PCR (normalized to miR-1). The y axis shows log2-tranformed values. Error bars are mean ± SD (n = 3). Bonferroni-corrected p value range: 2.8 × 10ˆ5 − 0.0049 (two-sample t tests).
(F) Scatterplots displaying each small RNA feature colored by class as a function of normalized log2-transformed small RNA reads in wild-type and mutant animals (axes are reverse transformed to reflect non-transformed values).
(G) Enrichment of rRNA-derived 22G-RNAs in FLAG::HRDE-1 co-immunoprecipitates relative to input cell lysates from wild-type and prg-1(n4357) mutants.
(H) Relative levels of a 26S/28S rRNA-derived 22G-RNA in wild-type and prg-1(tm872) mutant animals as determined by TaqMan qRT-PCR (normalized to miR-1). Error bars are mean ± SD (n = 3). The p value was calculated using a two-sample t test.
(I) Normalized log2-transformed rRNA-derived 22G-RNA levels in wild-type and prg-1(n4357) and mut-16(pk710) mutant gonads. Error bars are mean ± SD (n = 3). p values were calculated using two-sample t tests followed by Bonferroni correction for multiple comparisons. The y axis shows log2-transformed values.
(J) Pie chart displaying the proportion of all WAGO-class 22G-RNA reads from features classified as 22G-RNA hyperaccumulators (>250 reads on average and >5-fold increase in 22G-RNA levels in prg-1 mutants relative to wild-type) or non-hyperaccumulators. WAGO-class features were expanded to include the hyperaccumulators that are not classified as WAGO targets.
To confirm that rRNAs are misrouted into the endogenous RNAi pathway in the absence of piRNAs, we tested whether mutations in core components of the RNAi machinery suppressed hyperproduction of 22G-RNAs in prg-1 mutants. Mutations in mut-14 and smut-1 (that are partially redundant RNA helicases), mut-2, or mut-7 all suppressed 22G-RNA amplification from rRNAs in prg-1 mutants, as determined by small RNA high-throughput sequencing and TaqMan qRT-PCR (Figures 2E and 2F) (Chen et al., 2005; Ketting et al., 1999; Phillips et al., 2014; Tijsterman et al., 2002; Zhang et al., 2011). Furthermore, aberrant 22G-RNA production was lost from all hyperaccumulators in prg-1;mut-14 smut-1 mutants, demonstrating that in general aberrant 22G-RNAs are produced through the endogenous RNAi pathway (Figure 2F).
Low levels of 26S/28S rRNA-derived 22G-RNAs accumulated in mut-14 smut-1 mutants and in animals containing mutations in both prg-1 and any one of the mutators tested, albeit at reduced levels compared to wild-type (Figures 2E and 2F). It is therefore possible that rRNAs are normally targeted by both the CSR-1-class 22G-RNA pathway thought to be involved in licensing gene expression and the WAGO-class 22G-RNA pathway involved in RNAi. Consistent with this possibility, rRNA-derived 22G-RNAs were previously shown to bind both CSR-1 and WAGO-1 (Zhou et al., 2017). We also found that 26S/28S rRNA-derived 22G-RNAs were enriched in co-immunoprecipitates (coIPs) from the nuclear Argonaute HRDE-1, which is involved in transgenerational gene silencing (Figure 2G) (coIPs from Phillips et al. [2015]) (Buckley et al., 2012). Furthermore, 22G-RNA enrichment was higher in coIPs from prg-1 mutants than from wild-type animals (Figure 2G). 26S/28S rRNA-derived 22G-RNA levels were also elevated in a distinct and unrelated allele of prg-1, tm872, demonstrating that this phenomenon is not unique to the prg-1(n4357) deletion allele primarily used in this study (Figure 2H). These results suggest that in the absence of piRNAs, rRNAs are directed into the heritable gene silencing pathway involving HRDE-1 and the RNAi machinery.
In total, 5.8S-derived 22G-RNA levels were elevated ~5-fold and 18S- and 26S/28S-derived 22G-RNAs levels were elevated >30-fold in prg-1 mutants (Figure 2I). Despite these elevated levels, we were unable to identify a consistent impact on rRNA levels in distal gonads of prg-1 mutants using qRT-PCR (Figure S2A). We also did not observe a difference in read distribution across the different rRNA subunits using total RNA-seq from dissected gonads of prg-1 and wild-type animals; nor did we capture reads derived from the 45S precursor outside of the subunits (Figure S2B). Nevertheless, while this manuscript was under review, another study identified a positive correlation between rDNA copy number and generational fertility in prg-1 mutants (Wahba et al., 2021). It is possible that we did not detect an effect by qRT-PCR or total RNA-seq because of the sheer abundance of rRNAs, or because variation in rRNA levels limited our ability to detect small differences.
Aberrant 22G-RNAs produced from rRNAs, histones, and other annotated coding genes in prg-1 mutants accounted for ~45% of all WAGO-class 22G-RNA reads in prg-1 mutants, but only a very small fraction in wild-type animals (Figure 2J). This points to substantial repurposing of the RNAi machinery for aberrant 22G-RNA production in prg-1 mutants. Disabling a single class of primary small RNAs that trigger WAGO-class 22G-RNA production (i.e., ERGO-1-class 26G-RNAs) enhances exogenous RNAi, likely because of competition for shared factors, suggesting that RNAi functions at or near capacity (Billi et al., 2014). The dramatic shift in WAGO-class 22G-RNA production to rRNAs, histones, and other coding genes may have wide-ranging impacts on 22G-RNA production in the germlines of prg-1 mutants. That many genes lose 22G-RNAs while other genes gain 22G-RNAs in prg-1 mutants suggests that piRNAs have a broad role in orchestrating endogenous RNAi.
Variation in aberrant 22G-RNA production between prg-1 mutant strains
We observed that the levels of 22G-RNAs produced from hyperaccumulators in prg-1 mutants were typically very consistent between biological replicates from the same parental line (Tables S1 and S3). To determine if hyperaccumulation of 22G-RNAs also occurs uniformly between different strains of prg-1 mutants, we assessed by high-throughput sequencing 22G-RNA levels in two distinct partial deletion alleles of prg-1, both of which are presumably null: prg-1(n4357) and prg-1(tm872) (Figures 3A and 3B) (Das et al., 2008; Yigit et al., 2006). The pattern of widespread loss of 22G-RNAs from some genes and gain of 22G-RNAs from others was strikingly similar between the prg-1 mutant strains (Figures 3A and 3B; Table S4).
Figure 3. Stochasticity of 22G-RNA production in prg-1 mutants.
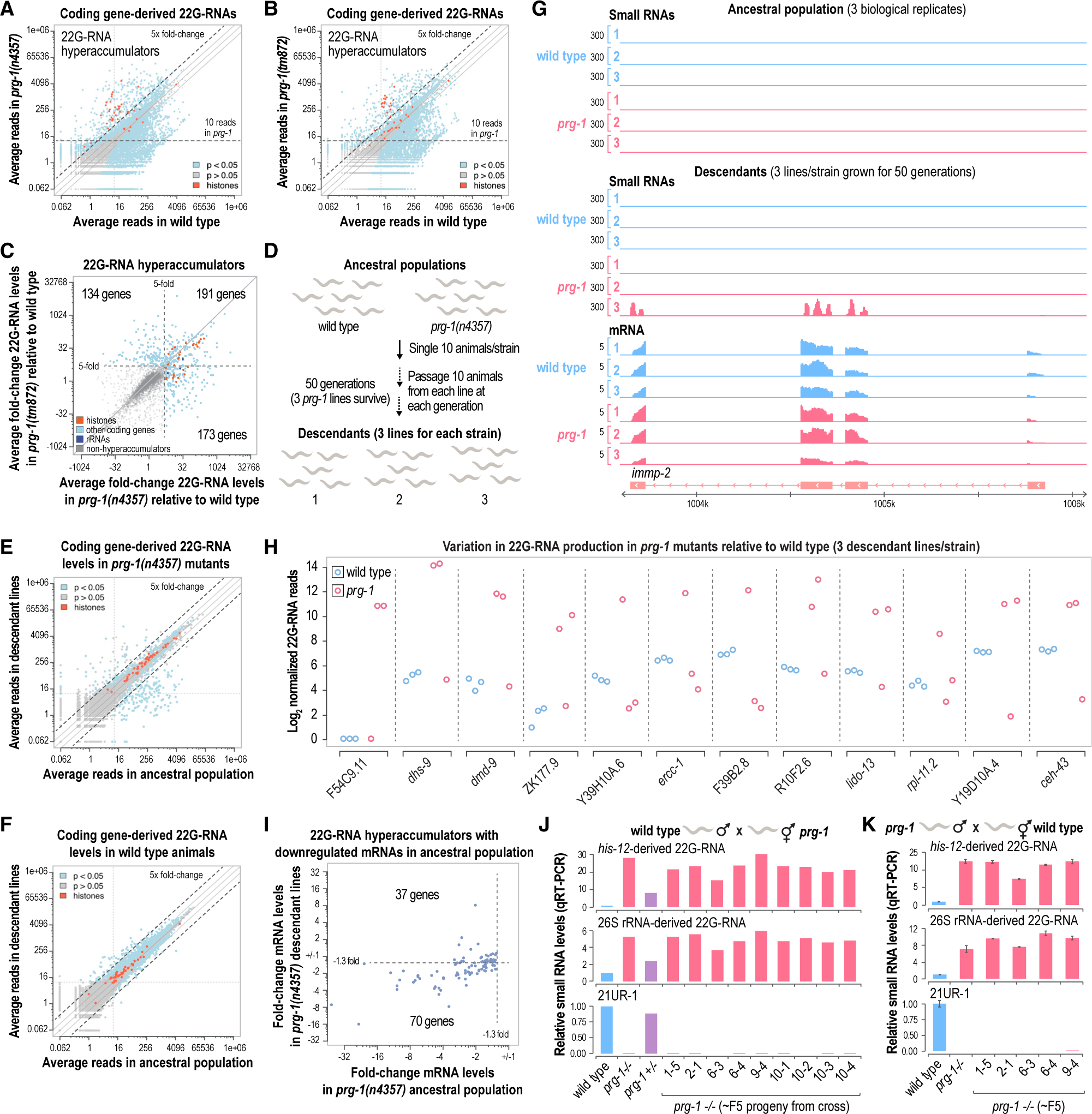
(A and B) Scatterplots displaying each gene as a function of normalized log2-transformed small RNA high-throughput sequencing reads in wild-type and prg-1(n4357) (A) or prg-1(tm872) (B) mutants (whole gravid adult animals, n = 3 biological replicates). The values on the axes are reverse transformed to reflect non-transformed values. Solid lines above and below the y = x lines indicate 2 and −2 fold-changes.
(C) Scatterplot displaying each gene classified as a 22G-RNA hyperaccumulator as a function of log2-transformed fold-change 22G-RNA levels in prg-1(n4357) or prg-1(tm872) relative to wild-type (whole gravid adult animals, n = 3). Genes that yielded >10 reads on average in at least one prg-1 mutant strain but that were not classified as hyperaccumulators are shown in gray. The axes reflect non-transformed values. The numbers of genes in each quadrant are shown. Dashed lines mark the 5-fold-change cutoff used to classify hyperaccumulators in the two prg-1 strains.
(D) Schematic illustrating the generational assay used in this study. mut-14(mg464) smut-1(tm1301) and prg-1(n4357); mut-14(mg464) smut-1(tm1301) strains were also included but for simplicity are not shown.
(E and F) Scatterplots displaying each gene as a function of normalized log2-transformed small RNA high-throughput sequencing reads in the ancestral population and descendant lines of prg-1(n4357) mutants (E) or wild-type animals (F) (whole gravid adult animals, n = 3). The axes show non-transformed values.
(G) mRNA and small RNA read distribution across a representative gene locus, immp-2. Three biological replicates or lines are shown for wild-type and prg-1(n4357).
(H) Plots displaying normalized log2-transformed small RNA reads for several genes in each wild-type and prg-1(n4357) mutant line after 50 generations of continuous growth (y axis shows log2-transformed values).
(I) Scatterplot displaying genes classified as 22G-RNA hyperaccumulators that were downregulated at the mRNA level as a function of mRNA fold-change in prg-1 mutants relative to wild-type at generations 1 (ancestral population) and 50 (descendant lines) of continuous growth.
(J and K) Bar plots displaying relative levels of each small RNA in the parental wild-type and prg-1 mutant lines and in the ~F5 progeny of crosses between wild-type males and prg-1(n4357) hermaphrodites (n = 1) (J) and prg-1(n4357) males and wild-type hermaphrodites (n = 2) (K). Error bars in (K) are mean ± SD.
Although we previously classified hyperaccumulators in prg-1(n4357), we repeated it for this experiment because it involved whole animals rather than dissected gonads. Because whole animals contain fewer 22G-RNAs relative to the total small RNA pool than gonad tissue (compare Figures 1B and 3A), we applied a less stringent, but arbitrary, 10 normalized read cutoff rather than the 250 read cutoff applied to our gonad-seq libraries for classifying hyperaccumulators. Using again a 5-fold-change cutoff to classify hyperaccumulators, we identified ~500 annotated genes that met our read threshold criteria in one or both strains (Table S4). Although some genes, including rRNAs and many histones, gained 22G-RNAs in both strains, many were unique to one of the two strains (Figure 3C; Table S4). There was also some variation in the extent to which genes lost 22G-RNAs in prg-1 mutants, however, the median fold change between the two prg-1 alleles among the hyperaccumulators was 5.9, compared with only 1.2 among non-hyperaccumulators (calculated from the data in Table S4). These results suggest that aberrant production of 22G-RNAs in prg-1 mutants, while seemingly consistent within a population derived from the same parental line, differs substantially between strains. Because many genes, such as histones and rRNAs, consistently gain 22G-RNAs in prg-1 mutants, there must be features that predispose some transcripts to 22G-RNA hyperproduction in the absence of piRNAs.
Stochasticity in 22G-RNA production within prg-1 populations
Having found that 22G-RNA production is variable between different strains of prg-1, we asked whether substantial variation also occurs within a population from a single strain. We first sequenced small RNAs and mRNAs from three distinct pools of wild-type animals and prg-1(n4357) mutants. The prg-1 strain was recently backcrossed to wild-type and then starved for an extended period. Starvation restores fertility to prg-1 mutants and brief bouts of starvation delay transgenerational sterility (Heestand et al., 2018; Simon et al., 2014). Thus, by starving the animals we were able to reset the fertility clock to some extent. The wild-type and prg-1 strains were then expanded on food for ~3 generations, after which, eggs were collected and hatched under starvation conditions to generate synchronized populations of L1 larvae, which were then transferred to food and grown to adult stage for RNA isolation. We refer to these founding populations as the ancestral generations, in which the animals were grown for 1 generation on continuous food. Next, we singled 10 animals from each population of wild-type and prg-1 mutant animals (as well as mut-14 smut-1 and prg-1; mut-14 smut-1, discussed below) and propagated them without starvation for 65 generations. Propagation across many generations in the presence of food leads to progressive germline dysfunction and loss of fertility in prg-1 mutants, which we also observed in this experiment (Heestand et al., 2018; Simon et al., 2014; Spichal et al., 2021).
At 50 generations, an arbitrary time point chosen because at this point the individual prg-1 lines had diverged considerably in health, as discussed below, we again sequenced small RNAs and mRNAs from the 3 surviving prg-1 mutant lines and 3 of the wild-type lines harvested as adults. We refer to these lines as descendants in reference to the ancestral populations from which they were derived (Figure 3D). Most genes that produced aberrant 22G-RNAs in the ancestral population of prg-1 mutants continued to do so in the descendant lines (Figures S3A and S3B). However, there was substantial variation in 22G-RNA levels between the ancestral population and the three descendent lines (Figure 3E). It is possible that 22G-RNA levels tend to drift across generations or that there is variation even within a population of wild-type animals. Arguing against this possibility, we did not observe dramatic gain or loss of 22G-RNAs between the ancestral population and the 3 descendant lines from wild-type animals (Figure 3F).
It is important to consider here that the descendant lines that were sequenced after 50 generations were pools of ~3,000 animals derived from similarly sized pools of animals that were sequenced at the ancestral generation. Within these pools there could be differences between individuals in which genes produced aberrant 22G-RNAs and thus we cannot assess with certainty gain or loss of 22G-RNAs within the three lines. Nevertheless, aberrant 22G-RNA production in only a subset of lines would point to stochasticity within the population. Indeed, we identified many genes for which only 1 or 2 of the 3 prg-1 descendant lines hyperaccumulated 22G-RNAs (Figures 3G and 3H; Table S5). In contrast, 22G-RNA levels were nearly identical in the 3 wild-type lines (Figure 3H; Table S5). In many instances, 22G-RNA levels were drastically different between the 3 prg-1 lines, indicating that aberrant 22G-RNA production is somewhat stochastic (Figure 3H; Table S5). However, we also observed drastic variation in the levels of 22G-RNA depletion in prg-1 mutants, and in some instances, 22G-RNA levels were reduced in one line and elevated in another (Figure S3C; Table S5).
We then did pairwise comparisons between each of the wild-type or prg-1 replicates to assess variation in coding gene-derived 22G-RNA levels more globally across the different lines. Within the ancestral populations, there was very little variation between the three biological replicates of either wild-type or prg-1 mutants (Figure S3D). In contrast, we observed dramatic variation in 22G-RNA levels between the three prg-1 descendant lines, which we did not observe between the wild-type descendant lines (Figure S3E). This variation was suppressed in prg-1; mut-14 smut-1 triple mutants, which were included in our generational assay as well, indicating that the variation we observed in the prg-1 single mutants is in WAGO-class, rather than CSR-1-class, 22G-RNA levels (Figure S3E).
The variation in 22G-RNA levels afforded us the ability to directly assess whether aberrant 22G-RNAs lead to silencing of the gene from which they are produced. The complexity of 22G-RNA production and mRNA expression somewhat obscured a global analysis. For example, many mRNAs were expressed at very low levels (Table S5). We did, however, identify several genes for which 22G-RNAs were clearly produced from an annotated transcript and mRNA levels were relatively high. In such cases, 22G-RNA levels were in some instances anticorrelated with mRNA levels (Figures 3G and S3F; Table S5). In many instances, however, there was no clear correlation between 22G-RNA levels and mRNA levels (Figure S3F; Table S5). These results, in combination with our earlier observation that many of the genes that produce aberrant 22G-RNAs in prg-1 mutants are downregulated (Figure 1D), indicate that piRNAs in some instances prevent aberrant gene silencing, but also further illustrate the inconsistent link between 22G-RNAs and gene silencing (Reed et al., 2020).
We then assessed whether genes that hyperaccumulated 22G-RNAs in prg-1 mutants in the ancestral generation and that were downregulated at the mRNA level were also downregulated in the descendant lines after 50 generations. Indeed, the majority of the 107 genes that were downregulated and hyperaccumulated 22G-RNAs in the ancestral lines were also downregulated in the descendant lines despite some stochasticity in 22G-RNA production and gene silencing among the three lines (Figure 3I; Table S5). We conclude that aberrant 22G-RNA production and gene silencing occur somewhat stochastically, but once initiated occur consistently across generations.
Aberrant 22G-RNA production from rRNAs and histones after outcrossing
We then asked whether the somewhat stochastic nature of aberrant 22G-RNA production we observed for many genes also applied to histones and rRNAs, as histones were consistently silenced in all our prg-1 mutant lines (Table S5). If so, outcrossing prg-1 mutants to wild-type might be able to restore normal levels of 22G-RNAs, at least partially. To test this, we did reciprocal crosses between wild-type and prg-1 mutant males and hermaphrodites. We then isolated multiple F1 progeny from the crosses and homozygosed and expanded them over a total of ~5–6 generations and then assessed aberrant 22G-RNA production from his-12 and 26S/28S rRNA using qRT-PCR.
In all prg-1 homozygous mutant lines, 22G-RNA levels were similar to those of the control prg-1(n4357) parental line regardless of whether the prg-1(n4357) mutation was introduced via the male or hermaphrodite, whereas a control heterozygous line showed an intermediate phenotype between wild-type and prg-1(n4357) mutants (Figures 3J and 3K). This suggests that aberrant 22G-RNA production from rRNAs and histones occurs consistently and within only a few generations in prg-1 mutants. Alternatively, aberrant 22G-RNAs or a memory of aberrant 22G-RNA production may be transmitted through both male and female gametes.
Aberrant gene silencing and generational fertility
Histone silencing was recently linked to the transgenerational sterility of prg-1 mutants (Barucci et al., 2020). Furthermore, our results demonstrating that rRNAs and many other coding genes are also susceptible to aberrant 22G-RNA production in prg-1 mutants led us to explore the link between aberrant gene silencing and transgenerational sterility in prg-1 mutants. During the experiments described above, we followed the fertility of prg-1 mutants, as well as mut-14 smut-1 mutants, which are defective in 22G-RNA production downstream of piRNAs, and triple prg-1; mut-14 smut-1 mutants over 65 generations (Figure 4A) (Phillips et al., 2014). Consistent with previous studies, most prg-1 mutant lines became sterile over the course of the experiment (Simon et al., 2014). Of the 10 prg-1 lines we followed, only 3 survived to generation 50 (Figure 4A). However, prg-1; mut-14 smut-1 mutants also displayed a high incidence of sterility (40%), which although lower than what was observed in prg-1 single mutants suggests that loss of WAGO-class 22G-RNAs, and thus presumably loss of aberrant gene silencing, cannot fully rescue the progressive sterility of prg-1 mutants (Figure 4A). Although, unexpectedly, mut-14 smut-1 mutants also displayed a progressive loss of fertility (Figure 4A).
Figure 4. Progressive sterility in piRNA- and 22G-RNA-deficient mutants.
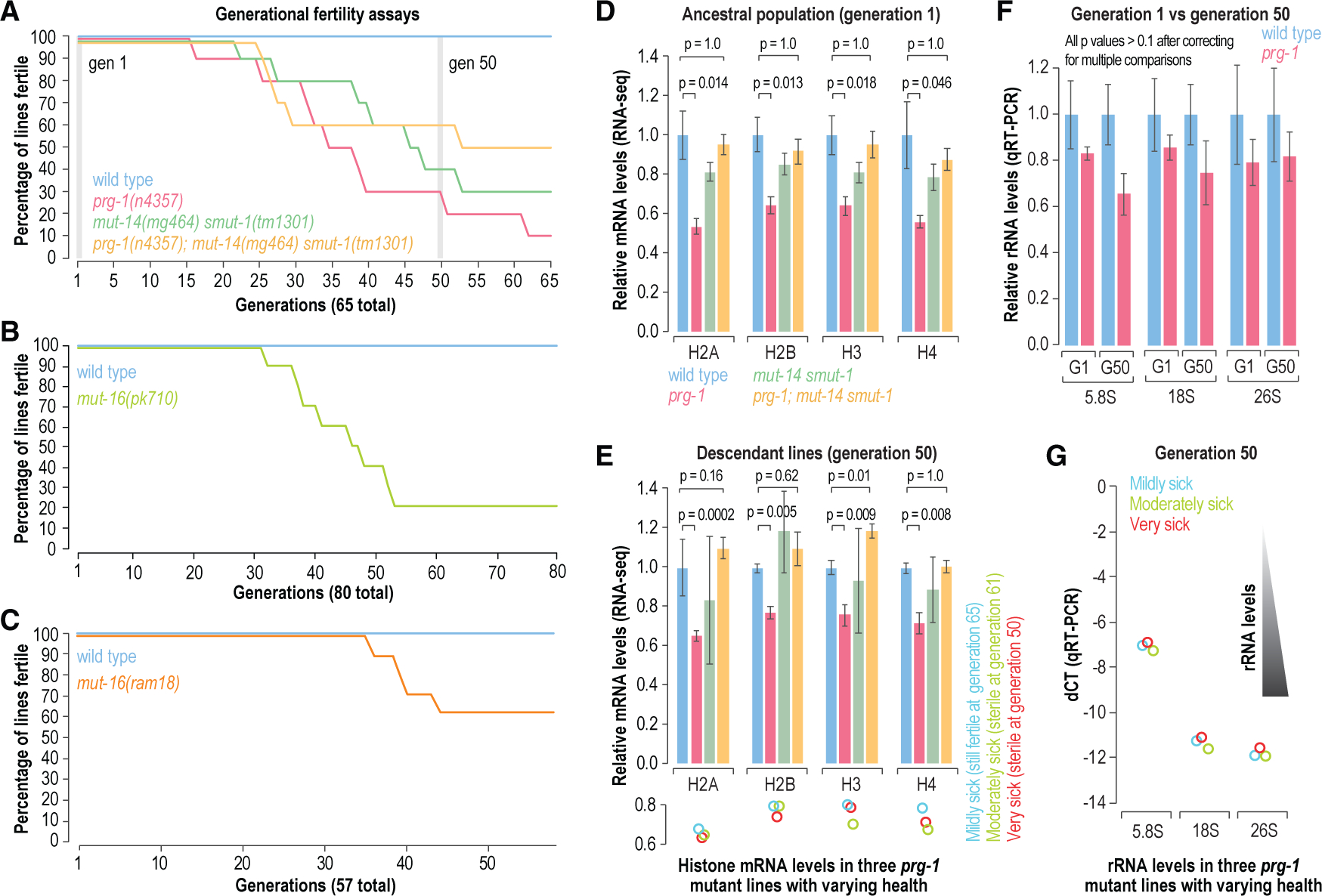
(A) Generational fertility of wild-type and prg-1(n4357), mut-14(mg464) smut-1(tm1301), and prg-1(n4357); mut-14(mg464) smut-1(tm1301) mutants. 10 lines for each strain were scored for fertility across 65 generations. The percentages of lines that were fertile at each generation are shown. RNA was isolated whole gravid adults from 3 lines per strain for RNA-seq at generations 1 (ancestral population) and 50 (descendant lines).
(B and C) Generational fertility of wild-type and mut-16(pk710) (B) or mut-16(ram18) (C) mutants. 10 lines for each strain were scored for fertility across 80 or 57 generations as indicated. The percentages of lines that were fertile at each generation are shown.
(D and E) Bar plots displaying relative mRNA levels for each histone family based on normalized RNA-seq counts after 1 (ancestral population) or 50 generations of continuous growth (descendant lines). Error bars are mean ± SD (n = 3). p values were calculated using two-sample t tests followed by Bonferroni correction for multiple comparisons. The lower circles-plot in (E) shows relative mRNA levels for each histone family in each of 3 prg-1(n4357) mutant lines colored by their health.
(F) Relative rRNA levels as determined by qRT-PCR in wild-type and prg-1(n4357) mutants at 1 and 50 generations of growth. Error bars are mean ± SD (n = 3). p values were calculated using two-sample t tests followed by Bonferroni correction for multiple comparisons.
(G) The circles-plot shows relative rRNA levels as determined by qRT-PCR for each rRNA family in each of 3 prg-1(n4357) mutant lines colored by their health. rRNA levels are represented by dCT ([mean cycle threshold for each rRNA – mean cycle threshold for actin] from 3 technical replicates). A 1 unit reduction in the dCT value would correspond to a 2-fold increase in rRNA levels.
To confirm that loss of WAGO-class 22G-RNAs underlies the progressive sterility of mut-14 smut-1 mutants, we assessed the fertility of two independent deletion alleles of mut-16, a gene also critical for WAGO-class 22G-RNA production, across 57 or 80 generations (Reed et al., 2020; Zhang et al., 2011). Lines from both mut-16 mutant strains also displayed generational sterility, although some lines persisted for >50 generations (Figures 4B and 4C). If aberrant gene silencing via 22G-RNAs is responsible for the progressive sterility of prg-1 mutants, it is surprising that 22G-RNA-defective animals also become sterile across generations. It is not clear, however, if the progressive sterility of mut-14 smut-1 and mut-16 mutants is related to the germline mortality observed in prg-1 mutants (Heestand et al., 2018; Simon et al., 2014; Spichal et al., 2021).
Finally, we explored the relationship between aberrant rRNA and histone silencing and transgenerational sterility. We observed almost no change in histone and rRNA 22G-RNA levels in prg-1 mutants across 50 generations of continuous growth, but it is possible that the ancestral lines were already producing 22G-RNAs at steady-state levels (Figure S3B). We nevertheless examined mRNA levels in prg-1 mutants at one generation of growth (the ancestral population) and in the 3 surviving lines at 50 generations of continuous growth (these are the same datasets for prg-1 and wild-type described above). Histone mRNA levels did not dramatically change in prg-1 single mutants between generation 1 and 50 but were rescued at both time points by the mut-14 smut-1 mutations, consistent with similar findings from Barucci et al. (2020) (Figures 4D and 4E).
Of the three prg-1 lines that were still viable at 50 generations, one was very sick and sterile, one was moderately sick and went sterile after 10 more generations, and one was only mildly sick and was fertile even at generation 65 when we terminated the experiment (Figure 4A). Despite disparity in their health, histone mRNAs levels in each of the lines were striking similar (Figure 4E, lower plot). We also assessed the levels of each of the rRNA families at 1 and 50 generations using qRT-PCR (because we selected against rRNA in our RNA-seq libraries). We were unable to detect a consistent difference in rRNA levels in prg-1 mutants at either time point and rRNA levels did not correlate with the health of the individual lines (Figures 4F and 4G). However, it is possible that the impact of reduced levels of histones or rRNAs is manifested over multiple generations without a further decline in their levels. Thus, despite a lack of supporting evidence, we cannot rule out that aberrant silencing of rRNAs or histones underlies the sterility of prg-1 mutants. Nevertheless, we propose that it may be a contributor rather than the underlying cause.
DISCUSSION
rRNAs and histone mRNAs are heavily targeted by PRG-1-piRNA complexes and yet it in the absence of piRNAs 22G-RNA production is upregulated rather than downregulated, suggesting that piRNAs somehow prevent, either directly or indirectly, overamplification of 22G-RNAs (Shen et al., 2018). It is possible that PRG-1-piRNA complexes physically block association with the 22G-RNA machinery to prevent 22G-RNA amplification under normal conditions. This hints at a possible mechanism for piRNA-mediated 22G-RNA formation: rather than physically directing mRNAs into the 22G-RNA pathway, piRNAs may act as a sieve of sorts that temporarily traps RNAs in germ granules. Presumably the trapped RNAs are acted on by gene-licensing factors, such as CSR-1, or gene-silencing factors, such as the WAGO Argonautes, or both. Some factors, such as WAGO or RdRP complexes, may be inefficient at displacing PRG-1-piRNA complexes from these trapped RNAs such that their ability to elicit an RNAi response is limited. The absence of PRG-1 could therefore enable access to the RNAi machinery and lead to runaway 22G-RNA amplification. This model may also explain why essentially all germline mRNAs are targeted by piRNAs, while only a subset is susceptible to RNAi (Reed et al., 2020; Shen et al., 2018; Wu et al., 2019). Perhaps a similar molecular sieve model could help to explain the role of the highly abundant pachytene piRNAs found in mice, which also have expansive gene targeting capacity (Ozata et al., 2019).
It is not clear why rRNAs and histone mRNAs are hypersusceptible to aberrant 22G-RNA production in the absence of piRNAs. Perhaps the sheer abundance of rRNAs and histone mRNAs overwhelms the cellular machinery that processes them. Without PRG-1 and polyA-binding proteins to help protect rRNAs and histones, 22G-RNAs may engage in a feedback loop that leads to runaway amplification by RNA-dependent RNA polymerases. It is possible that misprocessed RNAs can act as a trigger for RNAi-mediated gene silencing. Such misprocessing would presumably be more likely to occur the more abundant a transcript is and is possibly more likely for transcripts such as histones and rRNAs for which 3’ end formation does not involve polyadenylation. Interestingly, a critical step in priming an RNA for 22G-RNA biogenesis is the addition of a polyUG tail (Shukla et al., 2020). This so-called pUGylation occurs on resected or cleaved mRNAs, indicating that polyA tails are first removed (Shukla et al., 2020; Tsai et al., 2015). rRNAs and histone mRNAs may be primed for pUGylation because they already lack polyA tails. In support of this, a recent paper published while this manuscript was under review showed that polyUG tails are added to histone mRNAs in prg-1 mutants (Shukla et al., 2021). Furthermore, it was shown that long-term multigenerational RNAi occurs in a subset of prg-1 mutants from the same population, demonstrating that there is stochasticity in the exogenous RNAi response as well as in endogenous RNAi (Shukla et al., 2021). The stochastic nature of aberrant 22G-RNA production in prg-1 mutants may be the result of a small number of misprocessed transcripts arising in an individual and triggering runaway amplification that persists across generations. Perhaps piRNAs help to trap such misprocessed mRNAs in germ granules where they are acted on by the RNA decay machinery and in the absence of piRNAs they migrate into the cytoplasm where they may be more prone to trigger 22G-RNA biogenesis.
We showed that loss of mut-16 or mut-14 and smut-1 also leads to transgenerational sterility. Germline mortality was not observed in mut-7 or rde-2 mutants, which are also key components of the 22G-RNA machinery (Simon et al., 2014). This may reflect a more modest loss of 22G-RNAs in these mutants compared to mut-16 and mut-14 smut-1 mutants (Phillips et al., 2014). Whether the sterility of mut-16 and mut-14 and smut-1 is associated with their role downstream of piRNAs is unclear. It is possible that it is caused by something distinct, for example the accumulation of transposon-induced mutations, which likely occur at higher frequencies in mutator mutants than in piRNA mutants (Barucci et al., 2020; Reed et al., 2020; Wallis et al., 2019). That loss of both piRNAs and WAGO-class 22G-RNAs leads to a slower decline in fertility points to at least partially distinct and opposing roles for the two pathways in the germline and is consistent with aberrant gene silencing having an impact on the fitness of prg-1 mutants.
We previously showed that the presence of piRNAs or a 22G-RNA-based memory of piRNA activity is critical for fertility within a single generation (Phillips et al., 2015). Reestablishing WAGO-class 22G-RNA production in animals that lack piRNAs leads to immediate and highly penetrant sterility. Interestingly, sterility is accompanied by aberrant silencing of essential genes and widespread mis-sorting of mRNAs between the CSR-1 and WAGO pathways (de Albuquerque et al., 2015; Phillips et al., 2015). It is possible that the aberrant gene silencing we observed in this study is the same phenomenon but kept in check to a greater degree by a functional WAGO pathway and a memory of piRNA activity. Perhaps the progressive loss of fertility in piRNA mutants is a result of both desilencing of potentially harmful genes and aberrant silencing of beneficial genes. The stochastic nature of both gain and loss of 22G-RNAs in prg-1 mutants could explain why sterility ensues over differing numbers of generations for individuals derived from the same parental lines.
Limitations of study
We are not able to conclude whether aberrant gene silencing underlies the sterility of prg-1 mutants. Because prg-1 sterility is at least partially rescued by disabling the endogenous RNAi pathway, aberrant gene silencing is likely a contributing factor. However, a direct link between aberrant silencing of a particular gene or class of genes, such as histones or rRNAs, and a downstream molecular phenotype, such as widespread promotion or suppression of gene expression, is lacking. Thus, it will be important to identify how the various roles identified for piRNAs in regulating gene expression contribute to germline immortality in C. elegans. Although aberrant silencing of essential genes may indeed underlie generational sterility, we believe that additional genetic or biochemical evidence will be needed to validate current models (Barucci et al., 2020; de Albuquerque et al., 2015; Phillips et al., 2015; Wahba et al., 2021). The challenge in such studies will be to disentangle direct roles for piRNAs in regulating gene expression from indirect effects on gene expression associated with defects in germline development.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Taiowa Montgomery (tai.montgomery@colostate.edu).
Materials availability
All C. elegans strains generated in this study are available on request from the Lead Contact without restriction.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available. Accession numbers are listed in the Key Resources Table.
All original code has been deposited at GitHub and is publicly available as of the date of publication. DOIs are listed in the Key Resources Table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Escherichia coli: OP50 | Gary Ruvkun (Harvard Medical School) | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Trizol | Life Technologies | Cat# 15596018 |
| RNA 5′ polyphosphatase | Illumina | Cat# RP8092H |
| NEBNext 2X PCR Master Mix | New England Biolabs | Cat# M0541L |
| TURBO DNase | Life Technologies | Cat# AM2238 |
| TURBO DNA-free Kit | Thermo Fisher Scientific | Cat# AM1907 |
| SuperScript III Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18080044 |
| Applied Biosystems TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | Cat# 4444557 |
| Taqman MicroRNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat# 4366596 |
| RNA 5′ Pyrophosphohydrolase | New England Biolabs | Cat# M0356S |
| Acrylamide/Bis 19:1, 40% (w/v) solution | Thermo Fisher Scientific | Cat# AM9022 |
| AMPure XP Beads | Beckman Coulter | Cat# A63881 |
|
| ||
| Critical commercial assays | ||
|
| ||
| NEBNext Ultra II Directional RNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7760S |
| TruSeq Stranded Total RNA Library Prep Human/Mouse/Rat kit | Illumina | Cat# 20020596 |
| RNA Clean & Concentrator | Zymo Research | Cat# R1015 |
| Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat) | Illumina | Cat# MRZH11124 |
| NEBNext Small RNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7300S, E7580S |
| TURBO DNA-free Kit | Thermo Fisher Scientific | Cat# AM1907 |
| SuperScript III Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18080044 |
| Applied Biosystems TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | Cat# 4444557 |
| Taqman MicroRNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat# 4366596 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat# 1725121 |
| Custom TaqMan Gene Expression Assay: actin: probe_sequence1:CCTTACGGATATCAATGTCG | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: actin: fwd_sequence: CACGAGACTTCTTACAACTCCATCA | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: actin: rev_sequence: AGAACAGTGTTGGCGTACAAGT | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 5.8 s rRNA: probe_sequence1: AAGCGTCTGCAATTCG | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 5.8 s rRNA: fwd_sequence: AGCTTGCTGCGTTACTTACCA | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 5.8 s rRNA: rev_sequence: GCGTTCGAAATTTCACCACTCT | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 18S rRNA: probe_sequence1: CTACGGTCCACGAATTT | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 18S rRNA: fwd_sequence: TTCGTATCATTACGCGAGAGGTG | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 18S rRNA: rev_sequence: CGCTGTTGGGCGTCTCA | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 28S rRNA: probe_sequence1: TTAGTCGCTCAAAGACCG | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 28S rRNA: fwd_sequence: GCGGTAACGCATTCGAACTTG | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 28S rRNA: rev_sequence: GGAGTCCGTTCCTTAACTAGAGAAG | Thermo Fisher Scientific | Cat# 4331348 |
| TaqMan MicroRNA Assay: miR-1 Assay ID 000385 | Thermo Fisher Scientific | Cat# 4427975 |
| Custom TaqMan MicroRNA Assay (his-12 22G-RNA): Target Sequence: GTTACCGGCCAACTCGAGAACC | Thermo Fisher Scientific | Cat# 4427975 |
| Custom TaqMan MicroRNA Assay (28S rRNA 22G-RNA): Target Sequence: GAAGAAAACTCTAGCTCGGTCT | Thermo Fisher Scientific | Cat# 4427975 |
| Custom TaqMan MicroRNA Assay (21UR-1): Target Sequence: TGGTACGTACGTTAACCGTGC | Thermo Fisher Scientific | Cat# 4427975 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed NGS data | This Paper | GEO: GSE179811 |
| Raw and analyzed NGS data | Reed et al., 2020 | GEO: GSE141243 |
| Raw and analyzed NGS data | Svendsen et al., 2019 | GEO: GSE137734 |
|
| ||
| Experimental models: organisms/strains | ||
|
| ||
| C. elegans: Bristol Strain N2 | Caenorhabditis Genetics Center | Strain N2 |
| C. elegans: NL4415[henn-1(pk2295)] | Kamminga et al., 2012 | N/A |
| C. elegans: SX922 [prg-1(n4357)] | Caenorhabditis Genetics Center | Strain SX922 |
| C. elegans: NL1810[mut-16(pk710)] | Caenorhabditis Genetics Center | Strain NL1810 |
| C. elegans: GR1948[mut-14(mg464) smut-1(tm1301)] | Phillips et al., 2015 | N/A |
| C. elegans: GR2070[prg-1(n4357); mut-14(mg464) smut-1(tm1301)] | Phillips et al., 2015 | N/A |
| C. elegans: TAM24[mut-16(ram18[ko(302-4051])] | Reed et al., 2020 | N/A |
| C. elegans: WM161[prg-1(tm872)] | Caenorhabditis Genetics Center | Strain WM161 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primer for RT-PCR: 45S rRNA Forward: CAACTGGCAAGAGTAGTGAC | Integrated DNA Technologies | N/A |
| Primer for RT-PCR: 45S rRNA Reverse: CTCGTGAACAA4CGTCTACTG | Integrated DNA Technologies | N/A |
| Primer for RT-PCR: rpl-32 Forward: CATGAGTCCGACAGATACCG | Integrated DNA Technologies | N/A |
| Primer for RT-PCR: rpl-32 Reverse: ACGAAGCGGGTTCTTCTGTC | Integrated DNA Technologies | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| CASHX v. 2.3 | Fahlgren et al., 2009 | N/A |
| DESeq2 v. 1.30.1 | Love et al., 2014 | DOI: https://bioconductor.org/packages/DESeq2 |
| RSEM v. 1.3.0 | Li and Dewey, 2011 | https://deweylab.github.io/RSEM/ |
| Star v. 2.5.0a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Fastp v. 0.20.1 | Chen et al., 2018 | https://github.com/OpenGene/fastp |
| RNA-seq workflow | This Paper | DOI: https://zenodo.org/record/5641385 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains
The Key Resources Table contains all C. elegans strains used in this study. Animals were grown at 20°C on nematode growth medium and fed E. coli OP50. Experiments were done using predominantly hermaphrodite adult stage animals (68–72 hours post L1 synchronization or 96 hours post egg prep if not synchronized by starvation). No efforts were made to select for or against males.
METHOD DETAILS
Strain generation
prg-1(n4357) I; mut-7(pk720) III and mut-2(ne298) prg-1(n4357) I double mutants were generated by crossing prg-1(n4357) I males to mut-7(pk720) III or mut-2(ne298) I hermaphrodites.
RNA isolation
Animals were synchronized by bleach treatment and hatched in M9 until arrested as L1 larvae, plated, and grown to gravid adult stage (68–72 hours post L1 synchronization). To prevent starvation, the 50 generation descendant lines were immediately plated on food after bleach treatment and grown to gravid adult stage (96 hours). RNA isolation from gonads was done in Reed et al. (2020). Animals used for RNA isolation in this study were washed three times in M9 buffer, flash frozen in liquid nitrogen, and lysed in Trizol. RNA was isolated using two rounds of chloroform extraction followed by isopropanol precipitation and DNase treatment.
mRNA-seq and total RNA-seq libraries
mRNA-seq and total RNA-seq libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina as described (Reed et al., 2020), or the TruSeq Stranded Total RNA Library Prep Human/Mouse/Rat kit following the manufacturer’s protocol. For mRNA-seq libraries, rRNA was depleted using the Ribo-Zero rRNA Removal Kit prior to library prep (this step was omitted for total RNA-seq libraries). Samples were sequenced on an Illumina sequencer (NextSeq 500, High Output Kit, Single-End, 75 Cycles, or HiSeq, Paired-End, 150 Cycles).
mRNA-seq data analysis
Data analysis was done as described (Reed et al., 2020). Briefly, quality filtering was done with fastp (Chen et al., 2018). Reads were mapped to the C. elegans genome (Wormbase release WS230) using Star (Dobin et al., 2013). Reads aligning to each feature were counted using RSEM (Li and Dewey, 2011). Differential expression analysis was done using DESeq2 (Love et al., 2014). Plots were drawn in R, Excel, and IGV (Thorvaldsdóttir et al., 2013). See Table S6 for additional details.
sRNA-seq libraries
Small RNA libraries were prepared as described (Reed et al., 2020). Briefly, 16–30-nt RNAs were size selected on 17% denaturing polyacrylamide gels and treated with RNA polyphosphatase or RppH (Almeida et al., 2019b) to reduce 5′ di- and triphosphates to monophosphates to enable 5′ adaptor ligation. Sequencing libraries were prepared with the NEBNext Multiplex Small RNA Library Prep Set for Illumina. Libraries were size selected on 10% polyacrylamide gels and sequenced on an Illumina sequencer (NextSeq 500, High Output Kit, Single-End, 75 Cycles, or HiSeq, Paired-End, 150 Cycles).
sRNA-seq data analysis
Small RNA data analysis was done as described (Reed et al., 2020). Briefly, small RNAs were parsed from adapters, quality filtered, and mapped to the C. elegans genome (Wormbase release WS230) using CASHX v. 2.3 (Fahlgren et al., 2009). Reads from specific features were counted using custom Perl scripts. Multimapping reads were normalized by the number of map sites. Differentially expressed small RNAs were identified using DESeq2 (Love et al., 2014). Plots were drawn in R, Excel, and IGV (Thorvaldsdóttir et al., 2013). See Table S6 for additional details.
Generational fertility assays
Generational fertility assays were modeled after the approach used by Simon et al. (2014) and Ahmed and Hodgkin (2000). 10 independent lines for each strain were monitored over multiple generations. At each generation, population-level fertility was scored and 10 worms per line were transferred to new plates. Animals producing any number of offspring were considered fertile. If a line produced fewer than 10 progeny all progeny were transferred.
qRT-PCR
Quantitative real-time PCR (qRT-PCR) was done as described (Reed et al., 2020) using SYBR Green or custom TaqMan assays. For SYBR Green qRT-PCR, iTaq Universal SYBR Green Supermix (Bio-Rad) was used with primers recognizing 45S and rpl-32 (normalization). Custom TaqMan gene expression assays using TaqMan Fast Advanced Master Mix were used to detect 5.8S, 18S, and 26S/28S rRNAs, act-1 (normalization), and 22G-RNAs derived from his-12 and 26S/28S rRNAs as well as 21UR-1 and hsa-miR-1 (used for normalization). All assays included non-template controls and, when applicable, melting curve analyses were done. Average Ct values were calculated for 3–4 biological replicates from 3 technical replicate PCRs. Relative small RNA or rRNA levels were calculated using 2ˆ-ddCt (Livak and Schmittgen, 2001).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details of each experiment can be found in the figures and figure legends. A significance threshold was set at p < 0.05 after correcting for multiple comparisons. Benjamin-Hochberg corrected p values, calculated using the Wald test in DESeq2 (Love et al., 2014), are reported for differential expression analysis. Two-sample t tests were used when analyzing histone family mRNA levels and qRT-PCR results. Bonferroni corrections were applied to account for multiple comparisons. Pearson correlation coefficients were used to measure linear correlation between biological replicate RNA-seq datasets.
Supplementary Material
Highlights.
Piwi/PRG-1 prevents aberrant gene silencing in C. elegans
Aberrant gene silencing in piwi/prg-1 mutants can occur for at least 50 generations
rRNAs are misrouted into the endogenous RNAi pathway in piwi/prg-1 mutants
Both gain and loss of siRNAs are stochastic in piwi/prg-1 mutants
ACKNOWLEDGMENTS
High-throughput sequencing was done at Colorado State University by Mark Stenglein, Marylee Layton, and Mikaela Samsel and at Novogene. Thanks to Dustin Updike, Carolyn Phillips, René Ketting, and Germano Cecere for comments and suggestions. Some strains used in this study were provided by the CGC, which is funded by the NIH (P40 OD010440). This work was supported by the NIH (R35GM119775 to T.A.M. and T32GM132057 to K.J.R.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110101.
INCLUSION AND DIVERSITY
While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
REFERENCES
- Ahmed S, and Hodgkin J (2000). MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403, 159–164. [DOI] [PubMed] [Google Scholar]
- Almeida MV, Andrade-Navarro MA, and Ketting RF (2019a). Function and Evolution of Nematode RNAi Pathways. Noncoding RNA 5, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida MV, de Jesus Domingues AM, Lukas H, Mendez-Lago M, and Ketting RF (2019b). RppH can faithfully replace TAP to allow cloning of 5’-triphosphate carrying small RNAs. MethodsX 6, 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. (2012). piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150, 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, and Miska EA (2012). Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barucci G, Cornes E, Singh M, Li B, Ugolini M, Samolygo A, Didier C, Dingli F, Loew D, Quarato P, and Cecere G (2020). Small-RNA-mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat. Cell Biol. 22, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. (2008). PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, and Kim JK (2012). The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 8, e1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billi AC, Fischer SE, and Kim JK (2014). Endogenous RNAi pathways in C. elegans. WormBook, 1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, and Kennedy S (2012). A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AC, and Updike DL (2015). CSR-1 and P granules suppress sperm-specific transcription in the C. elegans germline. Development 142, 1745–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecere G, Hoersch S, O’Keeffe S, Sachidanandam R, and Grishok A (2014). Global effects of the CSR-1 RNA interference pathway on the transcriptional landscape. Nat. Struct. Mol. Biol. 21, 358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Simard MJ, Tabara H, Brownell DR, McCollough JA, and Mello CC (2005). A member of the polymerase beta nucleotidyltransferase superfamily is required for RNA interference in C. elegans. Curr. Biol. 15, 378–383. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, and Gu J (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb JM (2014). Ancient endo-siRNA pathways reveal new tricks. Curr. Biol. 24, R703–R715. [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. (2009). The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Moresco JJ, Gu W, Shirayama M, Conte D Jr., Yates JR 3rd, and Mello CC (2013). Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell 155, 1532–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. (2008). Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31, 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque BF, Placentino M, and Ketting RF (2015). Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans. Dev. Cell 34, 448–456. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Sullivan CM, Kasschau KD, Chapman EJ, Cumbie JS, Montgomery TA, Gilbert SD, Dasenko M, Backman TW, Givan SA, and Carrington JC (2009). Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA 15, 992–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson-Gurwitz A, Wang S, Sathe S, Green R, Yeo GW, Oegema K, and Desai A (2016). A Small RNA-Catalytic Argonaute Pathway Tunes Germline Transcript Levels to Ensure Embryonic Divisions. Cell 165, 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr., Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. (2009). Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol. Cell 36, 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heestand B, Simon M, Frenk S, Titov D, and Ahmed S (2018). Transgenerational Sterility of Piwi Mutants Represents a Dynamic Form of Adult Reproductive Diapause. Cell Rep. 23, 156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki YW, Siomi MC, and Siomi H (2015). PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 84, 405–433. [DOI] [PubMed] [Google Scholar]
- Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, and Ketting RF (2012). Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 8, e1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Haverkamp TH, van Luenen HG, and Plasterk RH (1999). Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Lee HC, Gu W, Shirayama M, Youngman E, Conte D Jr., and Mello CC (2012). C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, and Ketting RF (2012). Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 31, 3422–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, and Ruvkun G (2012). PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 8, e1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, and Zamore PD (2019). PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20, 89–108. [DOI] [PubMed] [Google Scholar]
- Pastore B, Hertz HL, Price IF, and Tang W (2021). pre-piRNA trimming and 2′-O-methylation protect piRNAs from 3′ tailing and degradation in C. elegans. Cell Rep. 36, 109640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Montgomery TA, Breen PC, and Ruvkun G (2012). MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Montgomery BE, Breen PC, Roovers EF, Rim YS, Ohsumi TK, Newman MA, van Wolfswinkel JC, Ketting RF, Ruvkun G, and Montgomery TA (2014). MUT-14 and SMUT-1 DEAD box RNA helicases have overlapping roles in germline RNAi and endogenous siRNA formation. Curr. Biol. 24, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CM, Brown KC, Montgomery BE, Ruvkun G, and Montgomery TA (2015). piRNAs and piRNA-Dependent siRNAs Protect Conserved and Essential C. elegans Genes from Misrouting into the RNAi Pathway. Dev. Cell 34, 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KJ, Svendsen JM, Brown KC, Montgomery BE, Marks TN, Vijayasarathy T, Parker DM, Nishimura EO, Updike DL, and Montgomery TA (2020). Widespread roles for piRNAs and WAGO-class siRNAs in shaping the germline transcriptome of Caenorhabditis elegans. Nucleic Acids Res. 48, 1811–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, and Bartel DP (2006). Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207. [DOI] [PubMed] [Google Scholar]
- Seth M, Shirayama M, Gu W, Ishidate T, Conte D Jr., and Mello CC (2013). The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell 27, 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen EZ, Chen H, Ozturk AR, Tu S, Shirayama M, Tang W, Ding YH, Dai SY, Weng Z, and Mello CC (2018). Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 172, 937–951.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D Jr., and Mello CC (2012). piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Yan J, Pagano DJ, Dodson AE, Fei Y, Gorham J, Seidman JG, Wickens M, and Kennedy S (2020). poly(UG)-tailed RNAs in genome protection and epigenetic inheritance. Nature 582, 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Perales R, and Kennedy S (2021). piRNAs coordinate poly(UG) tailing to prevent aberrant and perpetual gene silencing. Curr. Biol. 31, 4473–4485.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Sarkies P, Ikegami K, Doebley AL, Goldstein LD, Mitchell J, Sakaguchi A, Miska EA, and Ahmed S (2014). Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep. 7, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spichal M, Heestand B, Billmyre KK, Frenk S, Mello CC, and Ahmed S (2021). Germ granule dysfunction is a hallmark and mirror of Piwi mutant sterility. Nat. Commun. 12, 1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Reed KJ, Vijayasarathy T, Montgomery BE, Tucci RM, Brown KC, Marks TN, Nguyen DAH, Phillips CM, and Montgomery TA (2019). henn-1/HEN1 Promotes Germline Immortality in Caenorhabditis elegans. Cell Rep. 29, 3187–3199.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, Ketting RF, Okihara KL, Sijen T, and Plasterk RH (2002). RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science 295, 694–697. [DOI] [PubMed] [Google Scholar]
- Tsai HY, Chen CC, Conte D Jr., Moresco JJ, Chaves DA, Mitani S, Yates JR 3rd, Tsai MD, and Mello CC (2015). A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell 160, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Hansen L, and Fire AZ (2021). An essential role for the piRNA pathway in regulating the ribosomal RNA pool in C. elegans. Dev. Cell 56, 2295–2312.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis DC, Nguyen DAH, Uebel CJ, and Phillips CM (2019). Visualization and Quantification of Transposon Activity in Caenorhabditis elegans RNAi Pathway Mutants. G3 (Bethesda) 9, 3825–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, and Reinke V (2008). A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 18, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeles CJ, Wu MZ, and Claycomb JM (2013). Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev. Cell 27, 664–671. [DOI] [PubMed] [Google Scholar]
- Wu WS, Brown JS, Chen TT, Chu YH, Huang WC, Tu S, and Lee HC (2019). piRTarBase: a database of piRNA targeting sites and their roles in gene regulation. Nucleic Acids Res. 47 (D1), D181–D187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, and Mello CC (2006). Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127, 747–757. [DOI] [PubMed] [Google Scholar]
- Zhang C, Montgomery TA, Gabel HW, Fischer SE, Phillips CM, Fahlgren N, Sullivan CM, Carrington JC, and Ruvkun G (2011). mut-16 and other mutator class genes modulate 22G and 26G siRNA pathways in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108, 1201–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tu S, Stubna M, Wu WS, Huang WC, Weng Z, and Lee HC (2018). The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 359, 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Feng X, Mao H, Li M, Xu F, Hu K, and Guang S (2017). RdRP-synthesized antisense ribosomal siRNAs silence pre-rRNA via the nuclear RNAi pathway. Nat. Struct. Mol. Biol. 24, 258–269. [DOI] [PubMed] [Google Scholar]
- Zhu C, Yan Q, Weng C, Hou X, Mao H, Liu D, Feng X, and Guang S (2018). Erroneous ribosomal RNAs promote the generation of antisense ribosomal siRNA. Proc. Natl. Acad. Sci. USA 115, 10082–10087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available. Accession numbers are listed in the Key Resources Table.
All original code has been deposited at GitHub and is publicly available as of the date of publication. DOIs are listed in the Key Resources Table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Escherichia coli: OP50 | Gary Ruvkun (Harvard Medical School) | N/A |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Trizol | Life Technologies | Cat# 15596018 |
| RNA 5′ polyphosphatase | Illumina | Cat# RP8092H |
| NEBNext 2X PCR Master Mix | New England Biolabs | Cat# M0541L |
| TURBO DNase | Life Technologies | Cat# AM2238 |
| TURBO DNA-free Kit | Thermo Fisher Scientific | Cat# AM1907 |
| SuperScript III Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18080044 |
| Applied Biosystems TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | Cat# 4444557 |
| Taqman MicroRNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat# 4366596 |
| RNA 5′ Pyrophosphohydrolase | New England Biolabs | Cat# M0356S |
| Acrylamide/Bis 19:1, 40% (w/v) solution | Thermo Fisher Scientific | Cat# AM9022 |
| AMPure XP Beads | Beckman Coulter | Cat# A63881 |
|
| ||
| Critical commercial assays | ||
|
| ||
| NEBNext Ultra II Directional RNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7760S |
| TruSeq Stranded Total RNA Library Prep Human/Mouse/Rat kit | Illumina | Cat# 20020596 |
| RNA Clean & Concentrator | Zymo Research | Cat# R1015 |
| Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat) | Illumina | Cat# MRZH11124 |
| NEBNext Small RNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7300S, E7580S |
| TURBO DNA-free Kit | Thermo Fisher Scientific | Cat# AM1907 |
| SuperScript III Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18080044 |
| Applied Biosystems TaqMan Fast Advanced Master Mix | Thermo Fisher Scientific | Cat# 4444557 |
| Taqman MicroRNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat# 4366596 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat# 1725121 |
| Custom TaqMan Gene Expression Assay: actin: probe_sequence1:CCTTACGGATATCAATGTCG | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: actin: fwd_sequence: CACGAGACTTCTTACAACTCCATCA | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: actin: rev_sequence: AGAACAGTGTTGGCGTACAAGT | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 5.8 s rRNA: probe_sequence1: AAGCGTCTGCAATTCG | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 5.8 s rRNA: fwd_sequence: AGCTTGCTGCGTTACTTACCA | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 5.8 s rRNA: rev_sequence: GCGTTCGAAATTTCACCACTCT | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 18S rRNA: probe_sequence1: CTACGGTCCACGAATTT | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 18S rRNA: fwd_sequence: TTCGTATCATTACGCGAGAGGTG | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 18S rRNA: rev_sequence: CGCTGTTGGGCGTCTCA | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 28S rRNA: probe_sequence1: TTAGTCGCTCAAAGACCG | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 28S rRNA: fwd_sequence: GCGGTAACGCATTCGAACTTG | Thermo Fisher Scientific | Cat# 4331348 |
| Custom TaqMan Gene Expression Assay: 28S rRNA: rev_sequence: GGAGTCCGTTCCTTAACTAGAGAAG | Thermo Fisher Scientific | Cat# 4331348 |
| TaqMan MicroRNA Assay: miR-1 Assay ID 000385 | Thermo Fisher Scientific | Cat# 4427975 |
| Custom TaqMan MicroRNA Assay (his-12 22G-RNA): Target Sequence: GTTACCGGCCAACTCGAGAACC | Thermo Fisher Scientific | Cat# 4427975 |
| Custom TaqMan MicroRNA Assay (28S rRNA 22G-RNA): Target Sequence: GAAGAAAACTCTAGCTCGGTCT | Thermo Fisher Scientific | Cat# 4427975 |
| Custom TaqMan MicroRNA Assay (21UR-1): Target Sequence: TGGTACGTACGTTAACCGTGC | Thermo Fisher Scientific | Cat# 4427975 |
|
| ||
| Deposited data | ||
|
| ||
| Raw and analyzed NGS data | This Paper | GEO: GSE179811 |
| Raw and analyzed NGS data | Reed et al., 2020 | GEO: GSE141243 |
| Raw and analyzed NGS data | Svendsen et al., 2019 | GEO: GSE137734 |
|
| ||
| Experimental models: organisms/strains | ||
|
| ||
| C. elegans: Bristol Strain N2 | Caenorhabditis Genetics Center | Strain N2 |
| C. elegans: NL4415[henn-1(pk2295)] | Kamminga et al., 2012 | N/A |
| C. elegans: SX922 [prg-1(n4357)] | Caenorhabditis Genetics Center | Strain SX922 |
| C. elegans: NL1810[mut-16(pk710)] | Caenorhabditis Genetics Center | Strain NL1810 |
| C. elegans: GR1948[mut-14(mg464) smut-1(tm1301)] | Phillips et al., 2015 | N/A |
| C. elegans: GR2070[prg-1(n4357); mut-14(mg464) smut-1(tm1301)] | Phillips et al., 2015 | N/A |
| C. elegans: TAM24[mut-16(ram18[ko(302-4051])] | Reed et al., 2020 | N/A |
| C. elegans: WM161[prg-1(tm872)] | Caenorhabditis Genetics Center | Strain WM161 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Primer for RT-PCR: 45S rRNA Forward: CAACTGGCAAGAGTAGTGAC | Integrated DNA Technologies | N/A |
| Primer for RT-PCR: 45S rRNA Reverse: CTCGTGAACAA4CGTCTACTG | Integrated DNA Technologies | N/A |
| Primer for RT-PCR: rpl-32 Forward: CATGAGTCCGACAGATACCG | Integrated DNA Technologies | N/A |
| Primer for RT-PCR: rpl-32 Reverse: ACGAAGCGGGTTCTTCTGTC | Integrated DNA Technologies | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| CASHX v. 2.3 | Fahlgren et al., 2009 | N/A |
| DESeq2 v. 1.30.1 | Love et al., 2014 | DOI: https://bioconductor.org/packages/DESeq2 |
| RSEM v. 1.3.0 | Li and Dewey, 2011 | https://deweylab.github.io/RSEM/ |
| Star v. 2.5.0a | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Fastp v. 0.20.1 | Chen et al., 2018 | https://github.com/OpenGene/fastp |
| RNA-seq workflow | This Paper | DOI: https://zenodo.org/record/5641385 |


