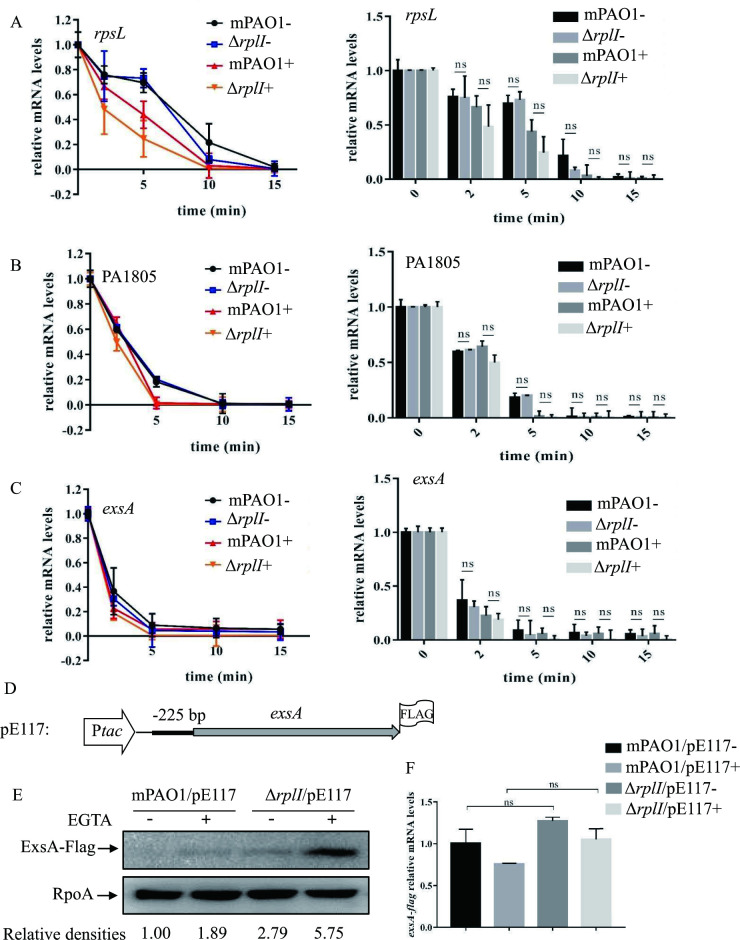
Fig 3. RplI controls the expression of ExsA at the posttranscriptional level by repressing ExsA translation.
(A-C) Degradation of rpsL (A), PA1805 (B), and exsA (C) mRNA in wild type mPAO1 and its ΔrplI mutant under T3SS inducing (+) and non-inducing (-) conditions with 5 mM EGTA. Bacterial cells were treated with rifampicin, collected at 2, 5, 10, and 15 min and mixed with equal numbers of gfp-expressing E. coli cells. Total RNA was purified, and the relative mRNA levels were determined by real-time qPCR. The gfp mRNA level in each sample was used as the internal control for normalization. The right panel is the bar diagram of the respective left panel. ns, not significant by Student’s t test. (D) Construct of exsA-Flag in pE117. (E) Amounts of ExsA-Flag. mPAO1 and ΔrplI containing pE117 were grown to an OD600 of 1.0 in LB with (+) or without (-) 5 mM EGTA and collected by centrifugation. Samples from equivalent numbers of bacterial cells were separated by SDS-PAGE and probed with an anti-Flag or an anti-RpoA antibody. The density of each band was determined with Image J. Relative densities represent the density of ExsA-Flag/density of RpoA with the first lane as 1. The data shown represent the results from three independent experiments. (F) Relative exsA-flag mRNA levels in mPAO1 and ΔrplI containing pE117. Total RNA was isolated under T3SS inducing (+) and non-inducing (-) conditions with 5 mM EGTA, and the exsA-flag mRNA levels were determined by real-time qPCR using rpsL as the internal control. ns, not significant, by Student’s t test.