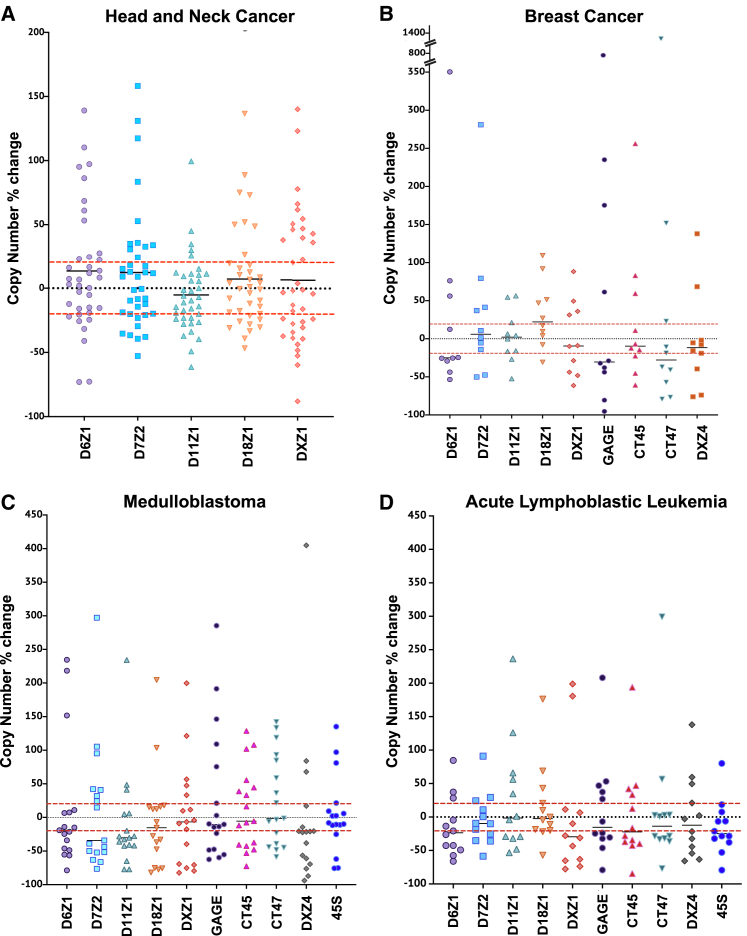
Figure 3.
Copy number changes in centromeric and tandemly repeated arrays in matched samples
Scatterplots contain points indicating the percentage of change from cancer, relative to normal, for each pair of samples. The absolute copy number difference was scaled to the size of the starting array of the normal tissue.
(A) The scatterplot depicts the percentage of change for the five indicated centromeric arrays in 35 head and neck cancer samples.
(B) The scatterplot depicts the percent change for five indicated centromeric and four tandemly repeated gene arrays (DXZ4, GAGE, CT45, and CT47) for 10 breast cancer samples.
(C) The scatterplot depicts the percentage of change for five indicated centromeric arrays and five tandemly repeated gene families (DXZ4, GAGE, CT45, CT47, and 45S rDNA) for 17 medulloblastoma samples.
(D) The scatterplot depicts the percentage of change for five indicated centromeric arrays in 12 acute lymphoblastic leukemia samples. The red dotted lines indicate the 20% window of error for the method; measurements within that range are considered “not significant.”