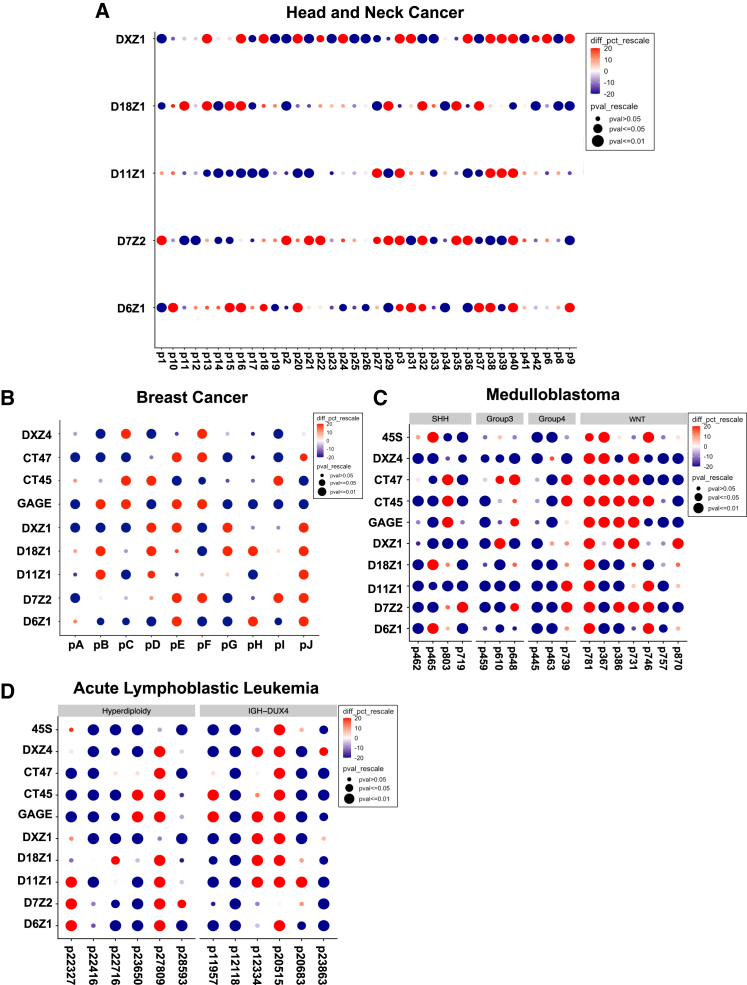
Figure 4.
Significant changes in arrays in each individual
Gridplots indicate the percentage and significance of changes per matched sample. Relative changes are scaled by the percentage of change (red is gain, and blue is loss) and p value (dot size).
(A) A gridplot is shown for the five indicated centromeric in 35 head and neck cancer samples.
(B) A gridplot is shown for five centromeric and four tandemly repeated gene arrays (DXZ4, GAGE, CT45, and CT47) for 10 breast cancer samples.
(C and D) A gridplot is shown for five indicated centromeric arrays and five tandemly repeated gene arrays (DXZ4, GAGE, CT45, CT47, and 45S rDNA) for 17 medulloblastoma samples (C) and 12 acute lymphoblastic leukemia samples (D), respectively. Medulloblastoma samples are categorized into four subgroups: WNT (n = 7), SHH (n = 4), group 3 (n = 3), and group 4 (n = 3), as previously described.28 85.7% (6/7) WNT samples have chromosome 6 loss. ALL samples are defined by hyperdiploidy (n = 6) or IGH-DUX4 (n = 6). High-hyperdiploid ALL samples exhibit non-random gain of chromosomes, which includes chromosomes 6, 10, 14, 21, and X. Gain of chromosomes 6, 18, and X was frequent in this study (see Table S4). Importantly, the methodology normalizes for chromosome copy number.