Abstract
Reverse transcription–loop-mediated isothermal amplification (RT-LAMP) has gained popularity for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The high specificity, sensitivity, simple protocols, and potential to deliver results without the use of expensive equipment has made it an attractive alternative to RT-PCR. However, the high cost per reaction, the centralized manufacturing of required reagents, and their distribution under cold chain shipping limit RT-LAMP’s applicability in low-income settings. The preparation of assays using homebrew enzymes and buffers has emerged worldwide as a response to these limitations and potential shortages. Here, we describe the production of Moloney murine leukemia virus reverse transcriptase and BstLF DNA polymerase for the local implementation of RT-LAMP reactions at low cost. These reagents compared favorably to commercial kits, and optimum concentrations were defined in order to reduce time to threshold, increase ON/OFF range, and minimize enzyme quantities per reaction. As a validation, we tested the performance of these reagents in the detection of SARS-CoV-2 from RNA extracted from clinical nasopharyngeal samples, obtaining high agreement between RT-LAMP and RT-PCR clinical results. The in-house preparation of these reactions results in an order of magnitude reduction in costs; thus, we provide protocols and DNA to enable the replication of these tests at other locations. These results contribute to the global effort of developing open and low-cost diagnostics that enable technological autonomy and distributed capacities in viral surveillance.
Keywords: SARS-CoV-2, diagnostics open source
INTRODUCTION
Coronavirus disease 2019 (COVID-19) is a respiratory infection caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).1, 2 The detection of SARS-CoV-2 has relied primarily on quantitative RT-PCR reaction.3, 4 Although quantitative RT-PCR is the gold-standard method for nucleic acid detection,5,6 this technique requires sophisticated equipment and skilled personnel, leading to centralized strategies of COVID-19 diagnosis with high turnaround times. These limitations have encouraged the search for alternative techniques for the detection of SARS-CoV-2. 7 Among them, loop-mediated isothermal amplification (LAMP) has gained popularity because of its high specificity, sensitivity, simple protocols, more tolerance to inhibitors, and potential to deliver results without the use of expensive instruments.8–10 These advantages make it ideal for decentralized testing and surveillance of human pathogenic agents (e.g., Dengue,11 Middle East respiratory syndrome,11 and HIV12, 13).
LAMP relies on DNA polymerases with strand displacement capability, primarily the large fragment (exo-) polymerase I from Geobacillus stearothermophilus (formerly Bacillus stearothermophilus), called Bst Large Fragment (BstLF).14 For RT-LAMP, the reaction is coupled with a reverse transcription step by adding a reverse transcriptase (e.g., HIV15 or the xenopolymerase RTx16) or utilizes a single enzyme with both activities (e.g., Bst3.017).
Despite these advantages, the high cost of enzymes can pose significant challenges to deployment in low-resourced areas. Enzymes are usually manufactured in a few centralized facilities located in the global north and distributed worldwide under cold chain shipping, which cannot always be maintained. Although there are no indications of RT-LAMP reagent shortages, centralized production models are prone to the global supply chain disruptions that have affected access to other diagnostic resources during the COVID-19 pandemic,18, 19 further limiting the testing capacity in many countries of the global south. In response to these limitations, many groups have described the successful implementation of homemade RT-PCR and RT-LAMP protocols based on homebrewed enzymes.15, 20–28
Here, we describe the in-house production of RT-LAMP reactions for SARS-CoV-2 from homebrewed Moloney murine leukemia virus (M-MLV) reverse transcriptase and BstLF polymerase. We compare their performance to commercial reactions and tested their ability to detect SARS-CoV-2 from RNA extracted from clinical nasopharyngeal samples.
MATERIALS AND METHODS
Expression and purification of M-MLV and BstLF
The M-MLV gene was synthesized as a Gblock from Integrated DNA Technologies (IDT) and cloned into a pET-19b vector with N-terminal 10× His-tag. BstLF DNA sequence from the OpenEnzyme collection was cloned into pET15b with a 6× His-tag at its N terminus. The BstLF sequence in the Open Enzyme collection was obtained from G. stearothermophilus (GenBank U23149, UniProt P52026). The large fragment was selected between the amino acids 298–876 of the full-length protein. The wild-type BstLF sequence was then codon optimized for Escherichia coli through the OPTIMIZER29 web server using the E. coli codon usage from the Highly Expressed Genes (HEG) database and a guided random method. Sequences were verified by Sanger sequencing services from Eton Bio, and full sequence files are provided here.30, 31 Both DNAs can be obtained from the Open Bioeconomy Lab and/or FreeGenes collection.32 M-MLV and BstLF were expressed in E. coli BL21(DE3) and C41, respectively, and purified following 2 open access protocols described in detail on the protocols.io Reagent Collaboration Network (ReClone) collection.33, 34 Briefly, protein production was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside for 16 h at 18°C at 200 rpm after plasmid-containing E. coli cells grown in ampicillinsupplemented lysogeny broth media reached OD600 = 0.8. Centrifugation-harvested cells were suspended in lysis buffer and disrupted first enzymatically using lysozyme and then with sonication on ice, using gentle cycles. Because BstLF is a thermostable protein, we added a thermolabile protein denaturation step by incubating the lysate at 65°C for 25 min. Lysates were then centrifuged at high speed to pellet cell debris, and the supernatants were recovered and purified on Nickel-nitrilotriacetic acid (Ni-NTA) pre-packed columns (HisTrap column, GE Healthcare). To remove imidazole, BstLF was buffer exchanged using Amicon Ultra-15 concentrators (Merck Millipore), whereas a second purification step using a HiTrap Heparin column (GE Healthcare) was employed for M-MLV. Protein concentration was determined by Bradford assay, using bovine serum albumin as standard.
RT-LAMP reaction setup
Buffers and reactions were prepared according to these online protocols (https://www.protocols.io/view/low-costlamp-and-rt-lamp-bsejnbcn) deposited in the ReClone collection in protocols.io.35 All reactions were performed on a StepOnePlus real-time thermocycler (Thermo Fisher Scientific, Waltham, MA, USA) using the SYBR green filter. Reactions with commercial enzymes Bst2.0 WarmStart [New England Biolabs (NEB), Ipswich, MA, USA]36 and RTx (NEB)36, 37 were performed according to the instructions from manufacturers. We used the primer set N238 purchased from IDT (https://www.idtdna.com) with standard desalting.
A fragment of nucleocapsid (N) gene RNA was used as positive control, which was produced by in vitro transcription from the IDT control for SARS-CoV-2 (catalog 10006625). A PCR amplicon was first obtained using the following primers: NT7_Fw: CGA AAT TAA TAC GAC TCA CTA TAG GGG CAA CGC GAT GAC GAT GGA TAG; T7_Nter_Rv: ACT GAT CAA AAA ACC CCT CAA GAC CCG TTT AGA GGC CCC AAG GGG TTA TGC TAG TTA GGC CTG AGT TGA GTC AG, which added a T7 promoter and terminator to the amplicon. In vitro transcription was next performed from this linear DNA using the Hi-Scribe kit from NEB (catalog E2040S) and treated with DNAse I (M0303S) for 15 min at 37°C before purification with Qiagen RNeasy kit, and serially diluted to the concentrations used.
The calculation of optimum enzyme concentrations was performed by maximizing an objective function that uses normalized functions for the amount of enzymes and the interpolation functions of normalized threshold time and amplification step size. A higher weighting was assigned to normalized threshold time and amplification step size to compute the optimal enzyme values. Further procedures and computational routines are detailed in our GitHub repository (https://github.com/LabTecLibres/RT-LAMP).
Clinical samples
RNA samples were obtained by RNA extraction from nasopharyngeal clinical samples previously collected from anonymous patients that attended the outpatient service of Red Salud UC-CHRISTUS (Santiago, Chile). RNA was extracted by automated machines MagNA Pure (Roche, Basel, Switzerland) or ExiPrep (Bioneer Corp, Daejeon, South Korea). Ct values of RT-PCR reactions were obtained with different primer sets for different samples, including United States Centers for Disease Control and Prevention (CDC) N1, N2, Orf1, and E sets. Samples with Ct values ≤ 40 were reported as positive SARS-CoV-2 cases. All methods were performed in accordance with the relevant guidelines and regulations. All procedures were approved by the Ethics Committee of Pontificia Universidad Católica de Chile.
Data and materials availability
All raw data are provided in a Zenodo repository (https://zenodo.org/record/4540817#.YCnTO1NKj8o). DNAs can be obtained upon request and also from the FreeGenes collection.32
RESULTS AND DISCUSSION
Expression and purification of M-MLV and BstLF
The M-MLV gene was commercially synthesized from a design that contains point mutations D200N, L603W, T330P, L139P, and E607K shown to increase thermostability and processivity.39 The large fragment was synthesized from a sequence design that was codon optimized for E. coli from amino acids 298–876 of the wild-type BstLF sequence from G. stearothermophilus (GenBank U23149, UniProt P52026).
BstLF and M-MLV were produced from isopropyl β-D-1-thiogalactopyranoside inducible plasmids pET15b BstLF and pET-19b_MMLV-RT bearing 6× and 10× His-tags at the N terminus, respectively, and expressed in E. coli, according to the open access protocols described in protocols.io.33, 40 Then, the purification was performed in one Ni-NTA chromatography step for BstLF and in 2 chromatography steps (Ni-NTA and Heparin) for M-MLV, resulting in highly purified proteins in 3–4 d. Typical protein purification yields from 1 L cell culture correspond to 4 mg of M-MLV and 10 mg of BstLF.
RT-LAMP reactions with homebrewed enzymes
To test the performance of the homebrewed enzymes, we compared RT-LAMP reactions composed of different combinations of BstLF, M-MLV, and commercial enzymes Bst2.0 (NEB) and RTx (NEB) in their suggested concentrations: 0.32 U/μl and 0.3 U/μl, respectively (Fig. 1A). We tested the amplification of 10,000 copies of an RNA fragment of N gene of SARS-CoV-2 using the N2 LAMP primer set.38 We found that 10.7 ng/μl of BstLF (and 62,5 pg/μl of M-MLV) to exhibit similar speed [i.e., time to threshold (Tt)] as the commercial enzymes (i.e., Bst2.0 + RTx), whereas 21.3 ng/μl and 5.3 ng/μl to be faster and slower, respectively. We also tested different buffers. First, we tested 2 commercial buffers, Isothermal Amplification Buffer Pack (IA buffer)41 and ThermoPol Reaction Buffer Pack (TPR Buffer),42 finding a better performance with IA buffer. Next, we compared homemade IA buffers with different concentrations of KCl: 50, 100, or 150 mM. Contrary to what was found in Aleeksenko et al.,22 we found that the IA buffer containing 50 mM KCl worked better than 100 and 150 mM (details in the online protocol). Mean Tt was 27.7 min for 50 mM of KCl, 43.5 min for 100 mM of KCl, and nondetectable for 150 mM of KCl. Therefore, homemade IA buffers containing 50 mM of KCl were used for the following assays.
FIGURE 1.
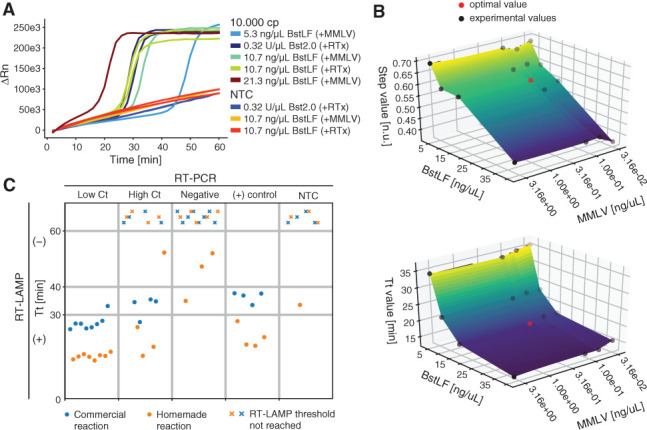
SARS-CoV-2 amplification with homemade RT-LAMP reactions. A) Comparison of RT-LAMP amplifications of 10,000 copies of an RNA fragment from gene Nusing N2 primer set and the following enzyme combinations: BstLF + M-MLV, BstLF + RTx (NEB), and Bst2.0 (NEB) + RTx (NEB). M-MLV was employed at 62.5 pg/μl in all reactions, whereas RTx was used at 0.3 U/μl, as indicated by the manufacturer. B) Enzyme concentration optimization according to Tt (top) and step (ON/OFF step; bottom). C) Detection of SARS-CoV-2 from RNA extracted from nasopharyngeal clinical samples. Positive controls correspond to 10,000 copies of a single-strand RNA (ssRNA) fragment from the N gene. “Threshold not reached” label corresponds to RT-LAMP reactions that did not amplify its target during the whole time of testing. For the homemade reactions, the mean Tt are 15 min for low Ct samples, 28 min for high Ct samples, 45 min for negative samples, and 21 min for positive samples. For the commercial reactions, the mean Tt are 27 min for low Ct samples, 33 min for high Ct samples, N/A for negative samples, and 36 min for positive samples. NTC, no template control.
Next, we studied the effect of different concentrations of enzymes on Tt and ON/OFF range (step) constrained by using the lowest number of enzymes possible in order to increase the number of reactions per purification (Fig. 1B). We found 16.52 ng/μl of BstLF and 0.127 ng/μl of M-MLV to be the optimum concentrations under these criteria. These concentrations were used for further assays and final tests with clinical samples.
Homebrew reaction tested with clinical samples
To address the potential use of in-house-prepared RT-LAMP reactions for the detection of SARS-CoV-2 from patient-purified RNA, we tested 16 positive samples (8 of high and 8 of low Ct RT-PCR values with primer set N1 and N2) and 8 negative samples with NEB WarmStart commercial kit and the homemade BstLF/M-MLV RT-LAMP reactions. The samples corresponded to RNA extracted from nasopharyngeal clinical samples (Fig. 1C). Although both reactions, commercial and homemade, were able to detect high viral load samples (i.e., low Ct values), negative samples tested with commercial enzymes showed no amplification in any tube (0/8) whereas homemade enzymes exhibited one false positive (1/7). Samples with low viral load (i.e., high Ct values), on the other hand, were more likely to be classified as negatives with both systems (4 out of 8 in commercial reactions and 3 out of 8 in homemade reactions). A careful analysis of melting curves showed that the positive results presented the expected curve for the target, indicating nonspurious amplification of the specific sequence (data not shown but provided in the Zenodo repository link with raw data). The reactions from negative clinical samples (i.e., negative RT-PCR reaction) were classified as negative with the commercial RT-LAMP system, whereas one tube was classified as positive in the homemade system. The melting curves showed that the positive result presented the expected curve of the target (i.e., not an amplification of spurious nonspecific sequences), as it was the case for the 2 RT-LAMP negative results with Tt values between 40 and 60 min, indicating a possible cross-contamination from other tubes.
Positive control reactions were correctly classified as positives in both systems. Negative controls were correctly classified with the commercial system, whereas the homemade reaction misclassified one tube as positive. The melting curves showed that the positive result presented the expected curve of the target (i.e., not an amplification of spurious nonspecific sequences), indicating a possible cross-contamination from another tube. Nevertheless, the Tt of false positives are clearly different from positive samples, indicating that a proper Tt selection (e.g., 30 min) should differentiate them. Although the analysis of melting curves suggests the absence of spurious unspecific amplifications, the homemade system provided faster times to threshold than the commercial reactions, which can be prone to deliver false negatives in reactions with high loads of nontarget RNAs, such as clinical samples.22 Nevertheless, these results demonstrate that RT-LAMP reactions composed of in-house purified BstLF and M-MLV have comparable performance to commercial kits. The costs of consumables for the homemade reactions (USD 0.36 per reaction) is one order of magnitude cheaper than the costs of reactions with commercial enzymes (NEB WarmStart kit, USD 3.86 per reaction, and NEB RTx and Bst2.0 WarmStart enzymes bought separately, USD 3.38 per reaction), enabling the development of more affordable alternatives for diagnostics. These results are in agreement with recent publications demonstrating the use of homebrewed enzymes for RT-LAMP detection of SARS-CoV-2 from patient samples.15, 22, 28 These efforts have demonstrated the use of in-house-produced BstLF and derivatives, version 7.16 and v5.9, along with a diverse set of homebrewed reverse transcriptases, such as RTx,28, 43 MashupRT (based on a feline leukemia virus reverse transcriptase FeLV-RT)22 and HIV RT.15 Here, we contribute with the use of inhouse-produced M-MLV for reverse transcription.
A variety of isothermal DNA amplification techniques have recently been developed and present themselves as highly versatile molecular biology tools to enable simple, in-the-field screening assays or point-of-care clinical diagnostics. Unlike traditional PCR, these reactions can be carried out at a constant temperature, eliminating the need for a thermocycler. Among these approaches is RT-LAMP, which has emerged as a promising and simple diagnostic tool for identifying SARS-CoV-2.9 Advances on detection methods for LAMP reactions have described a series of alternatives with different degrees of equipment dependency (or none), such as turbidity, gel electrophoresis, calcein, colorimetric indicators, pH-sensitive dyes, and DNA-intercalating fluorescent dyes (e.g., SYBR Green, SYTO dyes, EvaGreen).44 Our implemented protocol relies on the use of intercalating dye EvaGreen, as it has been shown to perform better than SYBR green.45, 46 A potential limitation of our proof-of-concept method is that the use of intercalating dyes can make target-specific real-time detection challenging44 and could pose as a barrier for use in resource-limited areas. Future lower-resource implementations of our method could substitute EvaGreen for 100–200 μM halochromic dyes, like Cresol Red, in pH 8.8 buffered reaction mix to cause detectable colorimetric change when amplification products have been produced.47 Alternatively, more advanced detection methods utilizing sequence-specific methods based on the use of probes (e.g., molecular beacons [LAMP-BEAC],23 oligonucleotide strand displacement [OSD],48 detection of amplification by releasing of quenching [DARQ],49 and quenching of unincorporated amplification signal reporters [QUASR]50) could also be utilized as end-point detection methods and remain to be tested in future work. Such methods would be advantageous, as they could be multiplexed for different targets in the same reaction. QUASR, for instance, has been shown to allow single-step and close-tube multiplexing.50
The versions of both M-MLV and BstLF enzymes used in this paper are in the public domain in Chile, but some of the M-MLV mutations are still subject to protection in other jurisdictions, e.g., in the United States (patent number US7056716B251 ), with anticipated expiration date of March 15, 2021. Future work will, therefore, explore the use of off-patent enzymes Open-MMLV or HIV RT15 from the E. coli expression toolkit from ReClone initiative52 in order to have freedom to operate in any country. The use of public-domain enzymes would also permit commercial partnerships seeking to scale up production locally. ReClone is a reagent collaboration network that addresses this challenge by providing access to open protocols and off-patent reagents for diagnostics under more permissive material transfer agreements (openMTA53), mitigating shortages as well as building preparedness in the long term. Overall, the Open RT-LAMP approach presented here demonstrates a viable method to replicate the detection sensitivities of traditional commercial RT-LAMP protocols at a fraction of the cost and with minimal resources. Although we demonstrate the utility of the Open RT-LAMP protocol in the detection of SARS-CoV-2 RNA isolated from clinical samples, this approach could easily be applied to a broad range of screening assays to identify other infectious diseases or common pathogens at low cost. Our Open RT-LAMP protocol aims to contribute to the global effort of making diagnostic capabilities more distributed, affordable, and equitable.
ACKNOWLEDGMENTS
This research was supported by ANID CONCYTEC covbio0012 and ANID – Millennium Science Initiative Program – ICN17_022. This work was further supported by the National Institutes of Health NIAID training grant (Training Program in Immunology; T32-AI07405) awarded to A.J.B. The authors thank the gLAMP consortium and the JOGL OpenCOVID community for the advice and for sharing relevant information for the establishment of LAMP reactions for SARS-CoV-2. The authors thank Carolina Miranda and Patricia Garcia from Red Salud UC-CHRISTUS for kindly providing PUC RNA samples from anonymous patients. The authors also thank Javier Gasulla (UBA, Argentina) for advice regarding the BstLF purification protocol. M.R. was funded by ANID FONDECYT 3190731 grant. P. Blázquez-Sánchez and J. Reyes were funded by ANID Doctoral Scholarship (ANID PFCHA 21191979 and 21191684, respectively). The authors declare no conflicts of interest.
REFERENCES
- 1.World Health Organization Naming the coronavirus disease (COVID-19) and the virus that causes it. Retrieved July 13, 2021, from https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it.
- 2.Zhu N, Zhang D, Wang W, et al. China Novel Coronavirus Investigating and Research Team A Novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med . 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang Y-W, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol . 2020;58:e00512–20. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Food and Drug Administration CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. Retrieved July 13, 2021, from https://www.fda.gov/media/134922/download.
- 5.Lu X, Wang L, Sakthivel SK et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis . 2020;26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esbin MN, Whitney ON, Chong S, Maurer A, Darzacq X, Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA . 2020;6:771–783. doi: 10.1261/rna.076232.120. (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guglielmi G. The explosion of new coronavirus tests that could help to end the pandemic. Nature . 2020;583:506–509. doi: 10.1038/d41586-020-02140-8. [DOI] [PubMed] [Google Scholar]
- 8.Thompson D, Lei Y. Mini review: recent progress in RT-LAMP enabled COVID-19 detection. Sensors and Actuators Reports . 2020;2:100017. doi: 10.1016/j.snr.2020.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Augustine R, Hasan A, Das S, Ahmed R, Mori Y, Notomi T et al. Loop-mediated isothermal amplification (LAMP): a rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology (Basel) . 2020;9:182. doi: 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teoh B-T, Sam SS, Tan KK, Johari J, Danlami MB, Hooi PS et al. Detection of dengue viruses using reverse transcription-loop-mediated isothermal amplification. BMC Infect Dis . 2013;13:387. doi: 10.1186/1471-2334-13-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosaka N, Ndembi N, Ishizaki A, Kageyama S, Numazaki K, Ichimura H. Rapid detection of human immunodeficiency virus type 1 group M by a reverse transcription-loop-mediated isothermal amplification assay. J Virol Methods. 2009;157:195–199. doi: 10.1016/j.jviromet.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtis KA, Rudolph DL, Owen SM. Rapid detection of HIV-1 by reverse-transcription, loop-mediated isothermal amplification (RT-LAMP) J Virol Methods . 2008;151:264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Kiefer JR, Mao C, Hansen CJ, Basehore SL, Hogrefe HH, Braman JC et al. Crystal structure of a thermostable Bacillus DNA polymerase I large fragment at 2.1 A resolution. Structure . 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 15.Kellner MJ, Ross JJ, Schnabl J bioRxiv . A rapid, highly sensitive and open-access SARS-CoV-2 detection assay for laboratory and home testing. 2020.06.23.166397 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv 2020.02.26.20028373 (2020). [DOI] [Google Scholar]
- 17.New England Biolabs Bst 3.0 DNA Polymerase. Retrieved July 13, 2021, from https://international.neb.com/products/m0374-bst-3-0-dna-polymerase#Product%20Information.
- 18.Nkengasong J. Let Africa into the market for COVID-19 diagnostics. Nature . 2020;580:565–565. doi: 10.1038/d41586-020-01265-0. [DOI] [PubMed] [Google Scholar]
- 19.Esfandiari S. Wuhan, China, faces such a shortage of coronavirus test-kits that people say getting one is like ‘winning the lottery’. Business Insider France Retrieved July 13, 2021, from https://www.businessinsider.fr/us/wuhan-coronavirus-china-shortage-test-kits-lottery-2020-1 (2020) [Google Scholar]
- 20.Maranhao A, Bhadra S, Paik I, Walker D, Ellington AD. An improved and readily available version of Bst DNA polymerase for LAMP, and applications to COVID-19 diagnostics. medRxiv . 2020.10.02.20203356 (2020) [Google Scholar]
- 21.Bhadra S, Maranhao AC, Ellington AD. One enzyme reverse transcription qPCR using Taq DNA polymerase. bioRxiv . doi: 10.1101/2020.05.27.120238. (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alekseenko A, Barrett D, Pareja-Sanchez Y, Howard RJ, Strand-back E, Ampah-Korsah H et al. Direct detection of SARS-CoV-2 using non-commercial RT-LAMP reagents on heat-inactivated samples. Sci Rep . 2021;11:1820. doi: 10.1038/s41598-020-80352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherrill-Mix S, et al. medRxiv . LAMP-BEAC: detection of SARS-CoV-2 RNA using RT-LAMP and molecular beacons. 2020.08.13.20173757 (2020) [DOI] [Google Scholar]
- 24.Bhadra S, Riedel TE, Lakhotia S, Tran ND, Ellington AD. bioRxiv . High-surety isothermal amplification and detection of SARS-CoV-2, including with crude enzymes. (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham TGW, et al. medRxiv . Inexpensive, versatile and open-source methods for SARS-CoV-2 detection. 2020.09.16. 20193466 (2020) [Google Scholar]
- 26.Mascuch SJ, Fakhretaha-Aval S, Bowman JC et al. A blueprint for academic laboratories to produce SARS-CoV-2 quantitative RT-PCR test kits. J Biol Chem . 2020;295:15438–15453. doi: 10.1074/jbc.RA120.015434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vonesch SC, et al. McQ – An open-source multiplexed SARS-CoV-2 quantification platform. medRxiv 2020.12.02.20242628 (2020) [Google Scholar]
- 28.Ali Z, Aman R, Mahas A, Rao GS et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res . 2020;288:198129. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puigbó P, Guzmán E, Romeu A, Garcia-Vallvé S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res . 2007;35(Web Server):W126–31. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.pET-19b_MMLV-RT Benchling Retrieved July 13, 2021, from https://benchling.com/s/seq-QR4HRtwI7IumNVoYKUeg.
- 31.pET15b_Bst-LF Benchling Retrieved July 13, 2021, from https://benchling.com/s/seq-5qNtNN9nE0HsQQLnX67N.
- 32.COVID-19 Diagnostic Toolkit Enzymes Retrieved July 13, 2021, from https://stanford.freegenes.org/products/covid-19-diagnostic-toolkit-enzymes. [Google Scholar]
- 33.Rivera M. Recombinant protein expression and purification of codon-optimized Bst-LF polymerase. 2020. [DOI]
- 34.Rivera M. 2020. Recombinant expression and purification of codonoptimized M-MLV and Mashup. [Google Scholar]
- 35.Molloy J. Reclone.org (The Reagent Collaboration Network) protocols.io Retrieved July 13, 2021, from https://www.protocols.io/groups/recloneorg-the-reagent-collaboration-network/recloneorg-the-reagent-collaboration-network/about (2020) [Google Scholar]
- 36.New England Biolabs Bst 2.0 WarmStart DNA Polymerase. Retrieved July 13, 2021, from https://international.neb.com/products/m0538-bst-20-warmstart-dna-polymerase#Product%20Information.
- 37.New England Biolabs WarmStart® RTx Reverse Transcriptase. Retrieved July 13, 2021, from https://international.neb.com/products/m0380-warmstart-rtx-reverse-transcriptase#Product%20Information.
- 38.Zhang Y, Ren G, Buss J, Barry AJ, Patton GC, Tanner NA. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques . 2020;69:178–185. doi: 10.2144/btn-2020-0078. [DOI] [PubMed] [Google Scholar]
- 39.Baranauskas A, Paliksa S, Alzbutas G, Vaitkevicius M, Lubiene J, Letukiene V et al. Generation and characterization of new highly thermostable and processive M-MuLV reverse transcriptase variants. Protein Eng Des Sel . 2012;25:657–668. doi: 10.1093/protein/gzs034. [DOI] [PubMed] [Google Scholar]
- 40.Brown A. Recombinant Protein Expression of MMLV-RT H (SkiBar H RT) V2 protocols.io. Retrieved July 13, 2021, from . [DOI]
- 41.Biolabs NE. Isothermal amplification buffer pack. Retrieved July 13, 2021, from https://international.neb.com/products/b0537-isothermal-amplification-buffer#Product%20Information. [Google Scholar]
- 42.New England Biolabs ThermoPol® Reaction Buffer Pack. Retrieved July 13, 2021, from https://international.neb.com/products/b9004-thermopol-reaction-buffer#Product%20Information. [Google Scholar]
- 43.Ellefson JW, Gollihar J, Shroff R, Shivram H, Iyer VR, Ellington AD. Synthetic evolutionary origin of a proofreading reverse transcriptase. Science. 2016;352:1590–1593. doi: 10.1126/science.aaf5409. [DOI] [PubMed] [Google Scholar]
- 44.Becherer L, et al. Loop-mediated isothermal amplification (LAMP) – review and classification of methods for sequence-specific detection. Anal Methods . 2020;12:717–746. [Google Scholar]
- 45.Lee SH, Baek YH, Kim YH, Choi YK, Song MS, Ahn JY. One-pot reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) for detecting MERS-CoV. Front Microbiol . 2017;7:2166. doi: 10.3389/fmicb.2016.02166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eischeid AC. SYTO dyes and EvaGreen outperform SYBR Green in real-time PCR. BMC Res Notes. 2011;4:263. doi: 10.1186/1756-0500-4-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanner NA, Zhang Y, Evans TC., Jr Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques . 2015;58:59–68. doi: 10.2144/000114253. [DOI] [PubMed] [Google Scholar]
- 48.Jiang YS, Bhadra S, Li B, Wu YR, Milligan JN, Ellington AD. Robust strand exchange reactions for the sequence-specific, real-time detection of nucleic acid amplicons. Anal Chem . 2015;87:3314–3320. doi: 10.1021/ac504387c. [DOI] [PubMed] [Google Scholar]
- 49.Tanner NA, Zhang Y, Evans TC. Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. BioTechniques . 2012;53(2):81–89. doi: 10.2144/0000113902. [DOI] [PubMed] [Google Scholar]
- 50.Ball CS, Light YK, Koh CY, Wheeler SS, Coffey LL, Meagher RJ. Quenching of Unincorporated Amplification Signal Reporters in Reverse-Transcription Loop-Mediated Isothermal Amplification Enabling Bright, Single-Step, Closed-Tube, and Multiplexed Detection of RNA Viruses. Analytical Chemistry . 2016;88(7):3562, 3568. doi: 10.1021/acs.analchem.5b04054. [DOI] [PubMed] [Google Scholar]
- 51.US 7056716 B2 - High fidelity reverse transcriptases and uses thereof the lens - free & open patent and scholarly search. Retrieved July 13, 2021, from https://www.lens.org/lens.
- 52.Reagent collaboration network – a global collaboration for equitable access to biotechnology. Retrieved July 13, 2021, from https://reclone.org.
- 53.Kahl L, Molloy J, Patron N, Matthewman C, Haseloff J, Grewal D et al. Opening options for material transfer. Nat Biotechnol . 2018;36:923–927. doi: 10.1038/nbt.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]


