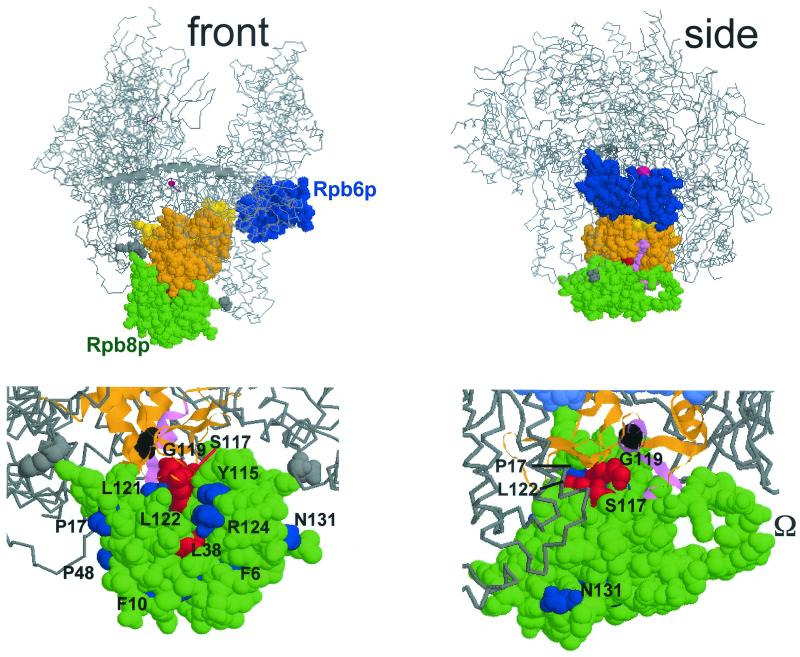
FIG. 4.
Spatial organization of Rpb8p and Rpb6p on Pol II. This figure was prepared with the RASMOL software (ftp://ftp.dcs.ed.ac.uk/pub /rasmol), using yeast Pol II coordinates (PDB accession codes, 1I3Q and 1I50; see reference 8). (Top) Front and side views approximately corresponding to the synomymous views used by Cramer et al. (8, 9). Rpb8p (green), Rpb6p (blue) without its first 71 amino acids, forming a disorganized tail (9, 11), and part of Rpb1p (between positions 508 and 663) are space-filled, the rest of the core Pol II structure being given as a Cα backbone, with the β'F bridge domain (positions 809 to 845 of Rpb1p) shown as an α-helix (see references 8 and 9 for more details on the spatial structure). Orange, two-hybrid interacting domain recognized by Rpb8p (positions 516 to 639; 13); violet, positions 563 to 580, corresponding to the invariant P.I.KP..LW.GKQ motif (see Fig. 3); yellow, part of the pore 1 module (positions 508 to 663; see reference 8) that is not included in the two-hybrid interacting domain; grey, K738 and D974, two positions that closely contact Rpb8p on the structure but are not included in the two-hybrid fragment. The catalytic Mg2+ is represented by a red sphere (not drawn to scale). (Bottom) Scaled-up views of the front and side views shown above, with slightly different viewing angles. The two-hybrid interacting domain is shown as an α-helical and β-stranded ribbon structure, with the same color code as above. Position G574 (corresponding to the rpa190-G728D mutation of Pol I; see Fig. 3) is space-filled and shown in black. Some of the Rpb8p positions mutagenized in this study are individualized in blue (phenotypically silent mutations) or red (lethal or conditional growth defects). The dispensable Ω loop (partly disorganized on the Rpb8p structure) (8, 21) is also indicated.