Extended Data Fig. 7. The human skin harbors vast viral diversity, of which the sebaceous sites remain stable over time.
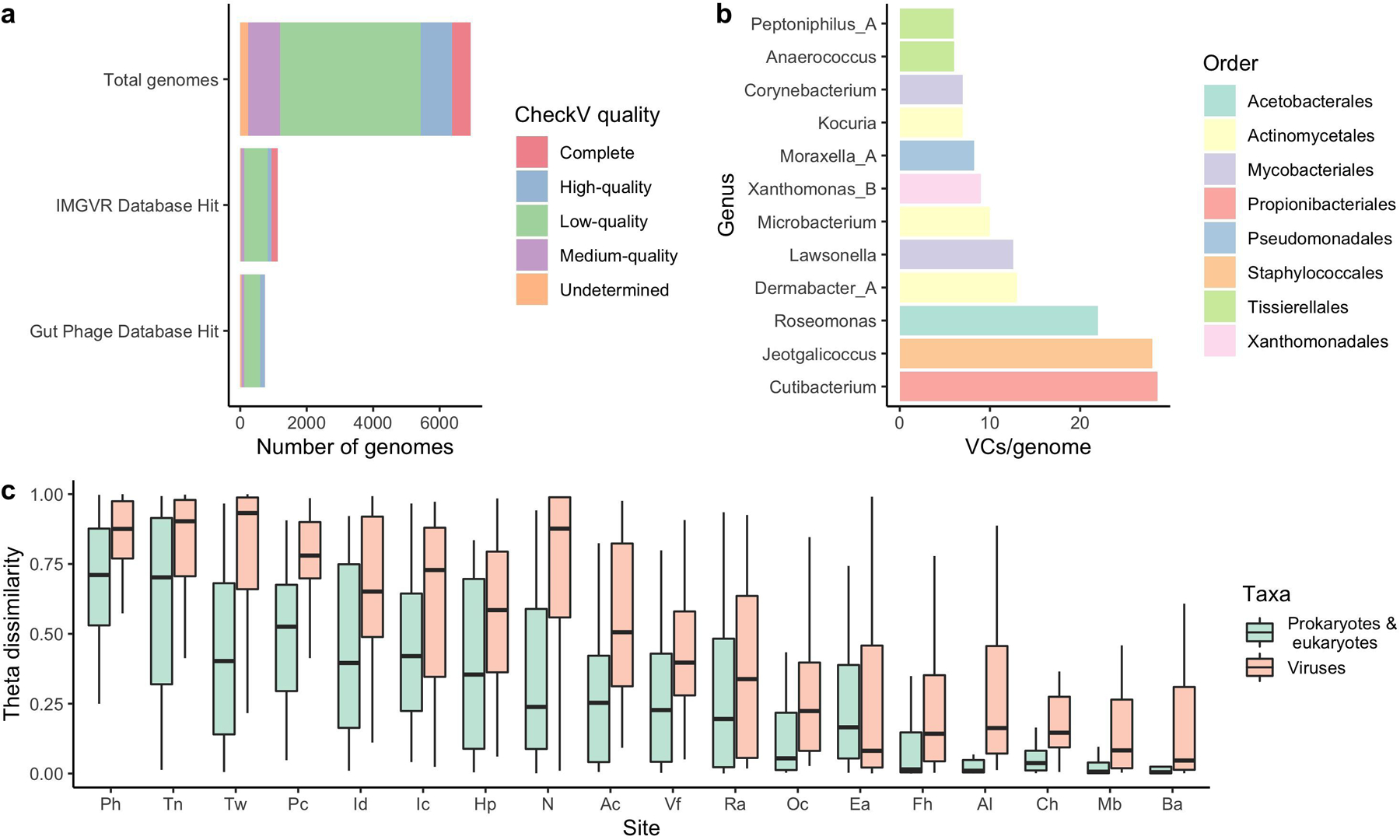
a, The number of viral genomes in the SMGC colored by their assigned CheckV quality. Comparison of the putative viral genomes to IMG/VR and the Gut Phage Database reveals that only a small fraction of the virome has been previously identified. b, The number of viral sequences detected for each SMGC bacterial genus using CRISPR host analysis. c, The stability of the SMGC over time for different body sites as estimated by the theta dissimilarity metric, with a theta dissimilarity of zero indicating high similarity. When calculating the theta dissimilarity, comparisons were made between the same body site of the same healthy volunteer over time. Body sites (Ac, n=39; Al, n=36; Ba, n=33; Ch, n=35; Ea, n=35; Fh, n=34; Hp, n=35; Ic, n=34; Id, n=32; Mb, n=35; N, n=42; Oc, n=36; Pc, n=35; Ph, n=36; Ra, n=41; Tn, n=32; Tw, n=35; Vf, n=38) are defined in Figure 1a. The Ax was excluded due to limited sampling. Box lengths represent the IQR of the data, with whiskers depicting the lowest and highest values within 1.5 times the IQR of the first and third quartiles, respectively.
