Extended Data Fig. 9. The SMGC improves classification of the skin microbiome.
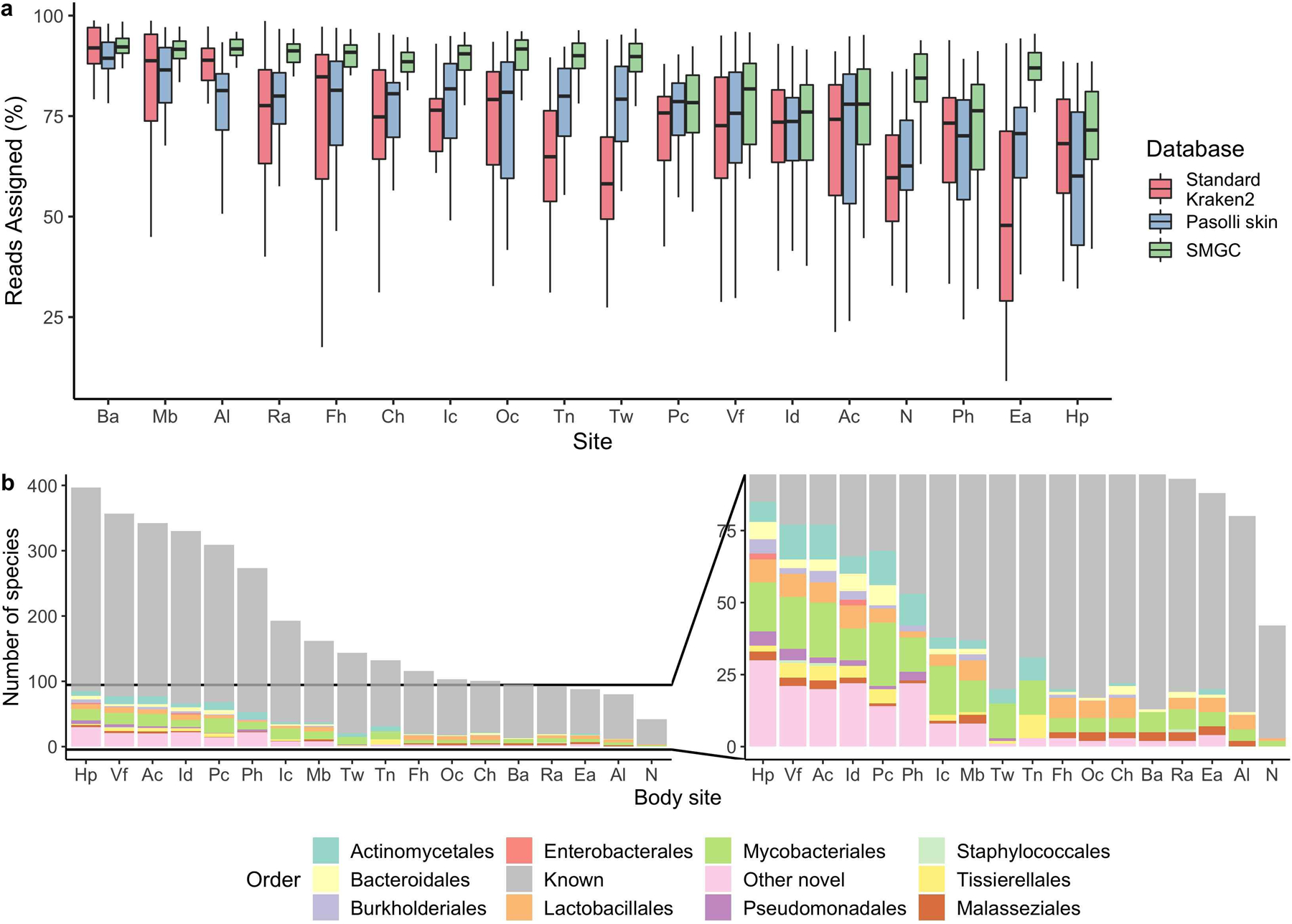
a, Percentage of sequencing reads from different body sites classified by the SMGC as compared to the standard Kraken 2 database and the Pasolli et al skin prokaryotic MAGs. Box lengths represent the IQR of the data, with whiskers depicting the lowest and highest values within 1.5 times the IQR of the first and third quartiles, respectively. b, The species in the SMGC present at different body sites. Novelty was determined by comparison to both the GTDB database and the Pasolli et al catalogue. Body sites (Ac, n=39; Al, n=36; Ba, n=33; Ch, n=35; Ea, n=35; Fh, n=34; Hp, n=35; Ic, n=34; Id, n=32; Mb, n=35; N, n=42; Oc, n=36; Pc, n=35; Ph, n=36; Ra, n=41; Tn, n=32; Tw, n=35; Vf, n=38) are defined in Figure 1a. The Ax was excluded due to limited sampling.
