Fig. 2. A comprehensive collection of skin microbial genomes uncovers abundant and prevalent bacterial diversity.
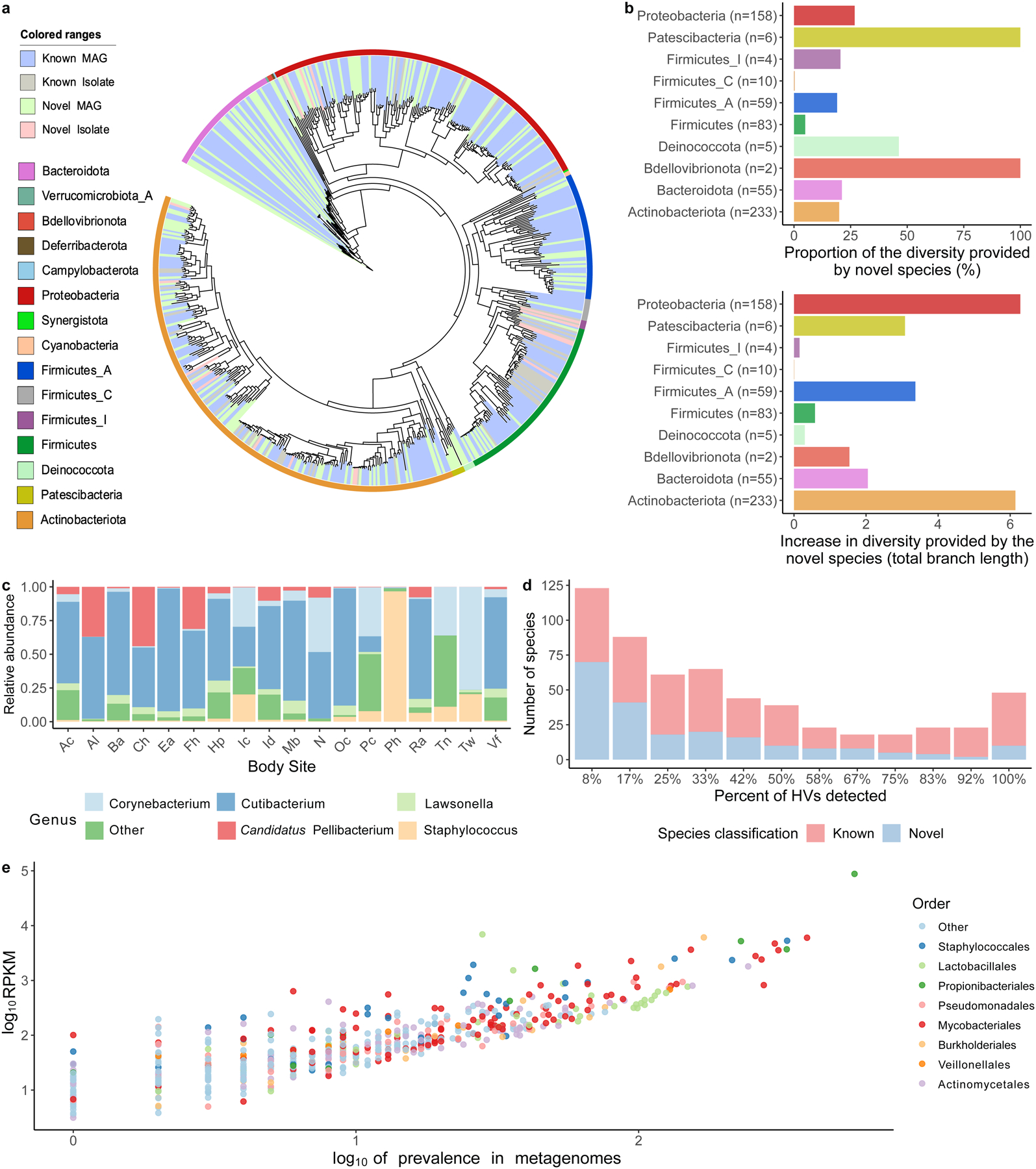
a, Phylogenetic tree of the 621 bacterial MAGs and cultured isolates colored by phyla and whether these are novel or known species. b, Level of phylogenetic diversity provided by the novel species relative to the complete diversity per phylum (top) and represented as absolute total branch lengths (bottom). The number of species from each phylum is depicted in brackets. c, Relative abundance of the most abundant genera found on the skin using the second time point of healthy volunteer HV03 as a representative. Body sites defined in Fig. 1a. d, The number of species shared between the 12 healthy volunteers, colored by their novelty. Presence was defined as having at least 30% of the bacterial genome covered in a sample from any time point or body site for a healthy volunteer. e, The prevalence and abundance (log10RPKM) of the SMGC colored by their taxonomic classification.
