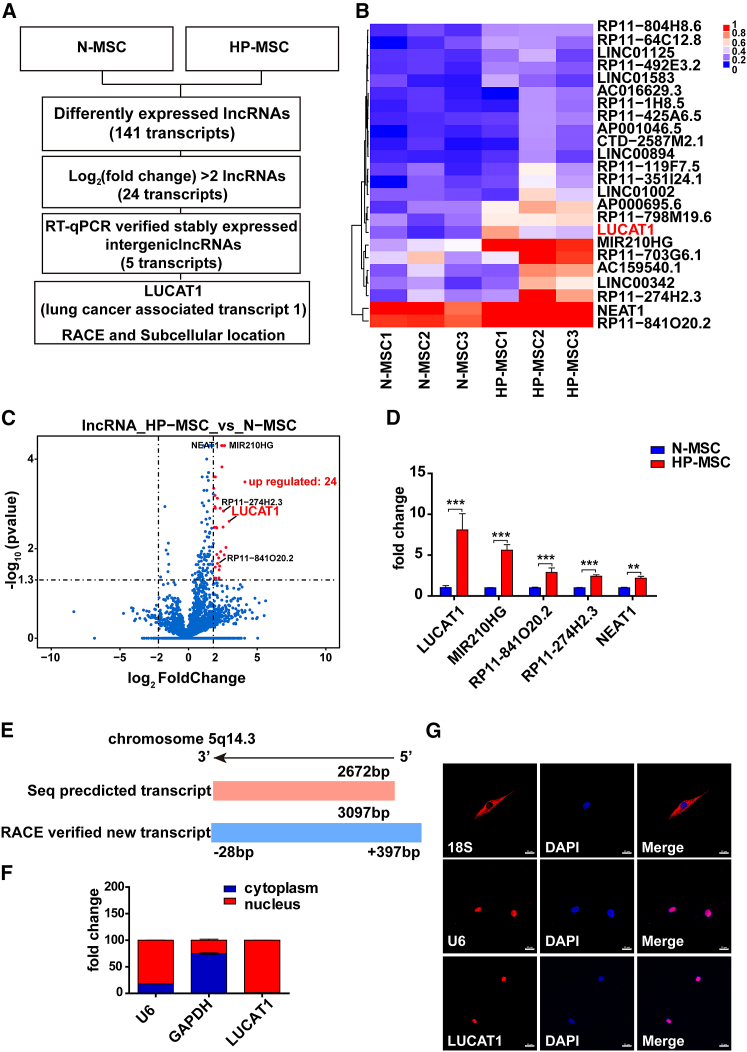
Figure 1.
LUCAT1 is highly expressed in HP-MSCs
(A) Schematic experimental design for screening specific lncRNAs. (B) lncRNA deep sequencing revealed long noncoding RNA expression profiles of HP-MSCs and N-MSCs from the three patient samples (fold change > 2; adjusted p value < 0.05). (C) The volcano map shows 24 differentially expressed lncRNAs (fold change > 2; adjusted p value < 0.05) in HP-MSCs, among which 5 lncRNAs showed upregulated expression in HP-MSCs. (D) qRT-PCR identified differentially expressed lncRNAs, with LUCAT1 having the highest fold change (n = 9). (E) RACE revealed that the genomic structure of LUCAT1-002 was a 3,097-nt transcript compared with the 2,627-nt transcript indexed in the Ensembl database. (F) Subcellular fractionation assays for evaluating the expression of LUCAT1 in MSC nuclei and cytoplasm (n = 3). (G) FISH for detecting the location of LUCAT1 in MSCs (bar, 20 μm). All data are presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.