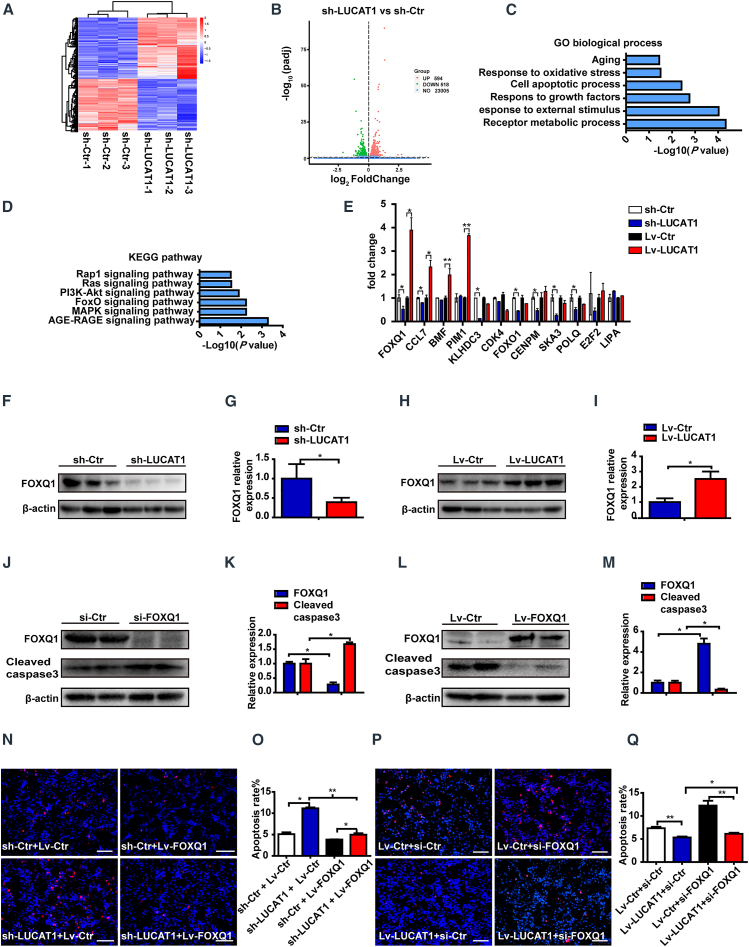
Figure 6.
LUCAT1 decreases MSC apoptosis through FOXQ1
(A) Heatmap of the LUCAT1-knockdown and control groups for 3 samples per group. (B) Volcano map revealed that 518 genes showed downregulated expression and 594 genes showed upregulated expression (adjusted p value < 0.05). (C) Gene Ontology (GO) terms for the protein-coding gene (PCG) cohort with downregulated expression in the LUCAT1-knockdown group. (D) Functional enrichment analysis was linked to biological pathways of the PCGs with downregulated expression in the LUCAT1-knockdown group. (E) The expression of relevant apoptosis-related genes was analyzed through qRT-PCR either for LUCAT1 knockdown or overexpression. (F–I) Western blot analysis for FOXQ1 in LUCAT1-knockdown (F and G) or -overexpression (H and I) MSCs (n = 3). (J–M) Cleaved caspase-3 levels after FOXQ1 knockdown (J and K) or overexpression (L and M) were assessed under 500-μM H2O2 for 1 h (n = 3). (N) Representative images of TUNEL staining with nuclei identified through DAPI staining in the control and LUCAT1-knockdown MSC groups with or without FOXQ1 overexpression under the above treatments (bar, 100 μm). (O) Apoptotic cells were quantified by TUNEL-positive nuclei in the total nuclei and analyzed via one-way ANOVA (n = 3). (P) Representative images of TUNEL staining with nuclei identified using DAPI staining in the control and LUCAT1-overexpression MSC groups with or without FOXQ1 knockdown as described above (bar, 100 μm). (Q) Apoptotic cells were quantified by TUNEL-positive nuclei in the total nuclei and analyzed via one-way ANOVA (n = 3). All data are presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.