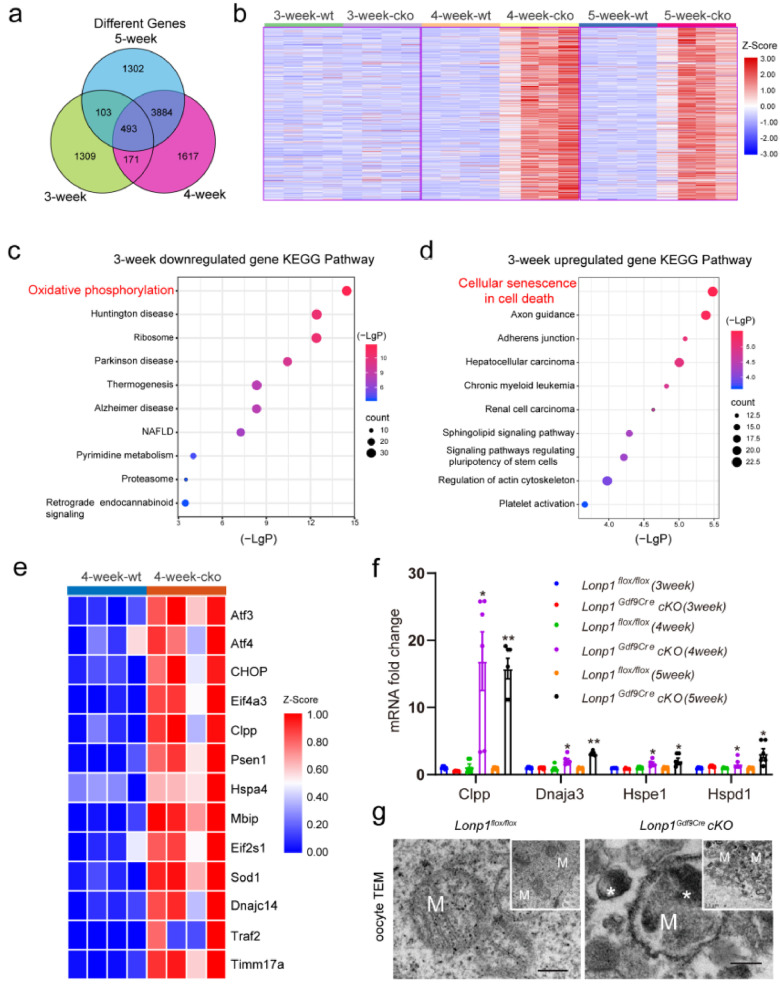
Figure 4.
Bioinformatics analysis showing prolonged mtUPR in Lonp1 knockout MII oocytes. (a) Venn diagram illustrating the relationships among the differentially expressed genes identified by RNA-seq analysis of MII oocytes from 3-, 4-, and 5-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=4 for each genotype). (b) SOM analysis of genes with altered transcript levels identified by RNA-seq analysis of MII oocytes from 3-, 4-, and 5-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=4 for each genotype). (c) KEGG enrichment analysis of significantly downregulated genes between MII oocytes from 3-week-old Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=4 for each genotype). (d) KEGG enrichment analysis of significantly upregulated genes between MII oocytes from 3-week-old Lonp1loxp/loxp mice and Lonp1Gdf9Cre cKO mice (n=4 for each genotype). (e) Differentially expressed genes involved in the mitochondrial stress response between MII oocytes from 4-week-old Lonp1loxp/loxp mice and Lonp1Gdf9Cre cKO mice (n=4 for each genotype). (f) qRT-PCR analysis of the expression of the mtUPR markers Clpp, Dnaja3, Hspe1 and Hspd1 in MII oocytes from Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=6, *p < 0.05, **p < 0.01). (g) Representative TEM images showing typical mitochondria in MII oocytes from Lonp1loxp/loxp and Lonp1Gdf9Cre cKO mice (n=3 for each genotype). The asterisks indicate high-electron-density protein accumulation. Scale bar=0.5 μm.