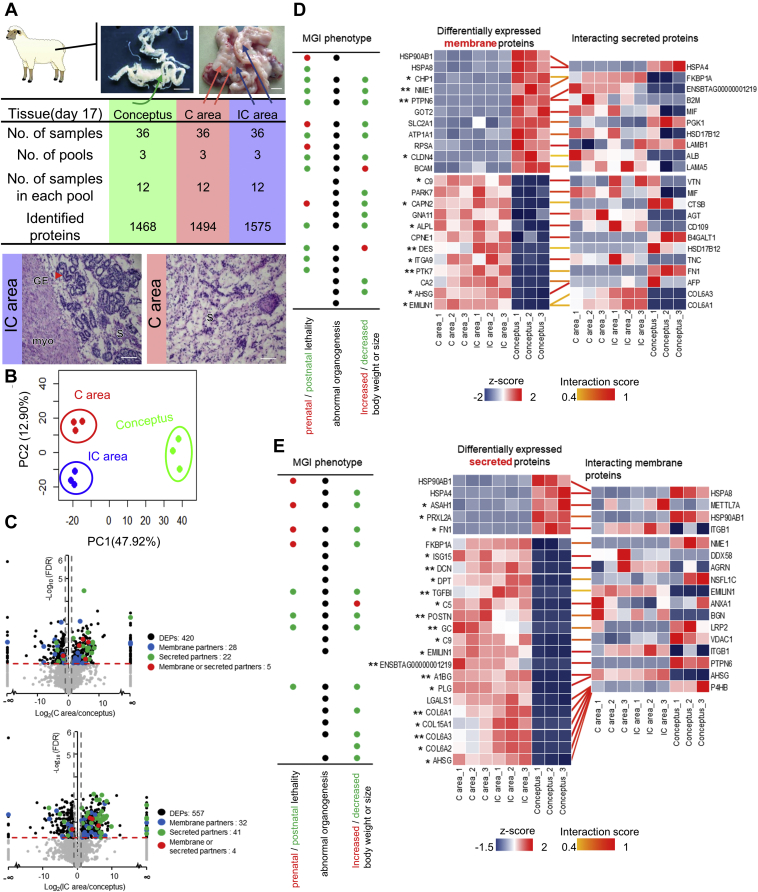
Figure 1.
Screening of differentially abundant membrane or secreted proteins and their interacting partners between the conceptus and endometrium.A, data structure used in this study. Representative images of a pregnant uterus and conceptus (scale bar = 1 cm). Data of genome-wide protein abundance were obtained from the conceptus, as well as the C and IC areas from 36 pregnant uteri, which were equally divided into three pools, 12 samples in each pool. Lower panel, representative hematoxylin-eosin staining images of C and IC areas identifying the tissues from which proteomic data were used in this study. Scale bar = 50 μm. B, principal component analysis of protein expression patterns of three biological replicates of the conceptus, C area, and IC area. C, volcano plot of differentially abundant proteins between the conceptus and the C area (upper panel) or IC area (lower panel). The black dots represent the differentially abundant proteins with FDR <0.05 and FC >2. The red line represents FDR = 0.05, and the gray line represents FC = 2. The blue dots represent identified membrane partners. The green dots represent identified secretion partners. The red dots represent proteins that could be secreted or located in membrane. D and E, overview of the interactions of membrane (D) or secreted (E) proteins that were significantly changed in both C and IC areas compared with that in the conceptus, with their interacting secreted/membrane partners. The Z-score normalized protein abundance is represented in red (relatively high) or blue (relatively low). ∗ represents the fold change of the protein in the conceptus versus that in the C areas and in the conceptus versus that in the IC areas, both >10. ∗∗ represents the fold change >100. The interaction scores are represented in yellow (0.4) to red (1). Left panel, the representative Mouse Genome Informatics (MGI) phenotypic annotations of differentially abundant membrane proteins. FDR, false discovery rate; GE, glandular epithelium; myo, myometrium; S, stroma.