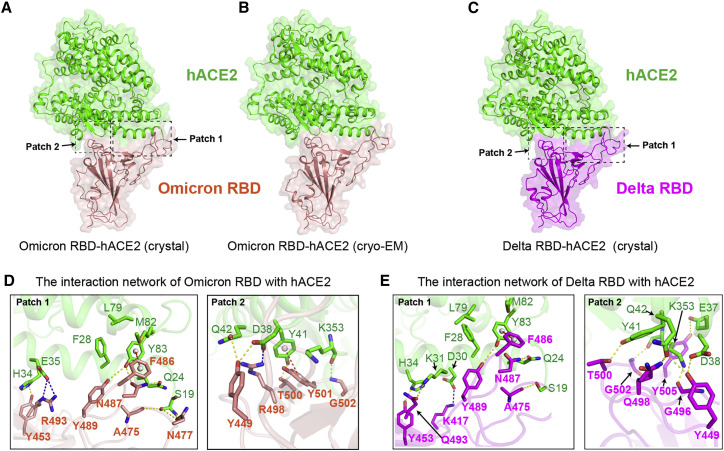
Figure 3.
Overall architecture and interaction network of hACE2-omicron RBD and hACE2-delta RBD complexes
(A–C) Overall architecture of hACE2-omicron RBD crystal structure (A) and cryo-EM structure (B) and crystal structure of hACE2-delta RBD complex (C).
(D) Interaction patches in crystal structure of hACE2-omicron RBD. Side chains of interacting residues on hACE2 (green) and omicron RBD (pink) are shown as sticks and labeled appropriately. Yellow, red, and blue dashes present H bond, π-π stacking interaction, and salt bridges, respectively.
(E) Interaction patches in crystal structure of hACE2-delta RBD. Side chains of interacting residues on hACE2 (green) and omicron RBD (purple) are shown as sticks. Yellow, red, and blue dashes present H bond, π-π stacking interaction, and salt bridges, respectively.
See also Figures S3, S4, and S5.