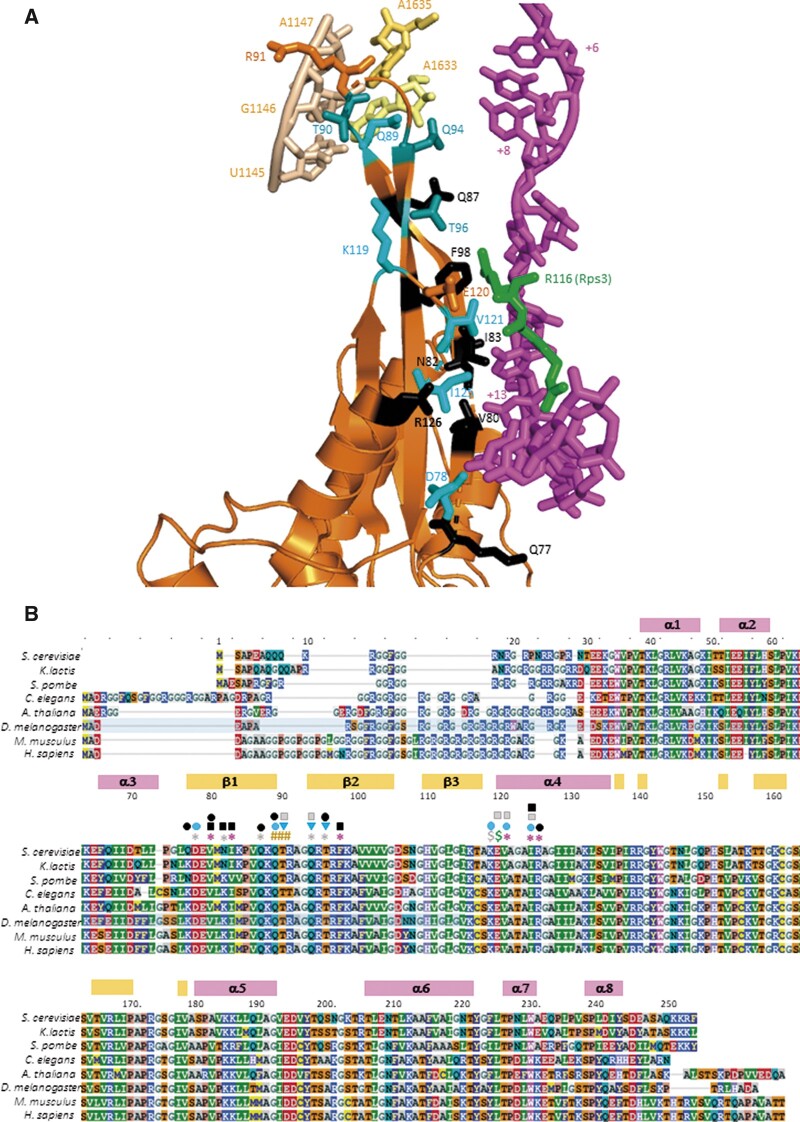
Figure 1.
Conserved uS5/Rps2 residues in proximity to the mRNA or rRNA at the 40S mRNA entry channel. (A) Cartoon depiction of the segment of Rps2 at the entry channel and its interactions with mRNA or rRNA constructed in PyMol from a partial yeast 48S complex in closed conformation (PDB: 6FYY), showing selected rRNA residues in h28 or the h28–h44 linker (wheat or pale yellow), mRNA nucleotides from +6 to +17 relative to the AUG (+1) (magenta), Rps3-R116 (green), and selected Rps2 residues (orange, cyan, or black). Side-chains are shown for Rps2 residues subjected to mutagenesis and colored black if only lethal substitutions were identified, light cyan if nonlethal substitutions were identified that increase discrimination against near-cognate UUG codons, and dark cyan (T90, Q94, and T96) for nonlethal substitutions that exclusively increase discrimination against start codons in poor Kozak context. (B) Multiple sequence alignment of Rps2 from the indicated species, with residues colored according to the chemical properties of the side chains, with boundaries and numbering of α-helices and β-strands indicated above the residue numbering for the S. cerevisiae sequence. Residues are annotated above the sequence as follows: red asterisks, 4–5Å from mRNA; gray asterisks, 6–8Å from mRNA; #, 4–5Å from rRNA; green $, 4 Å from Rps3-R116; gray $, 6 Å from Rps3-R116; black squares, recessive lethal; black ovals, dominant lethal; gray squares, Slg−; cyan ovals, strong Ssu−; cyan triangles, weak Ssu−.