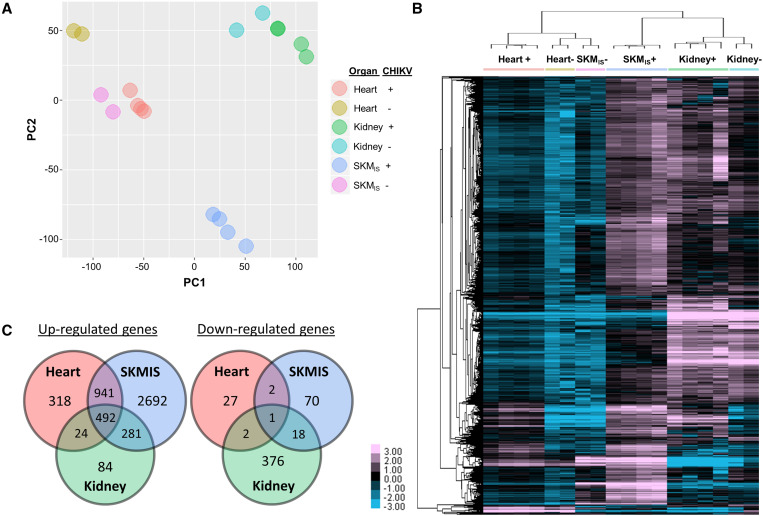
Figure 2.
Differential gene expression profiles for tissues from Chikungunya virus (CHIKV)-infected A129 mice. Poly-A primed ClickSeq libraries were constructed with RNA samples collected from chikungunya virus-infected (CHIKV; N = 4, noted by “+”) or mock-infected (N = 2, noted by “−”) A129 mouse kidney, skeletal muscle (SKMIS), and heart to evaluate changes in gene expression. Significant (fold change ≥ 2, P-adj ≤ 0.1) gene expression changes were identified with DPAC pipeline. (A) Principal component analysis showed clear consistency among replicates and distinctive clustering by both tissue and CHIKV infection status. (B) Hierarchical clustering of the significantly altered genes using Cluster 3.0 followed by TreeView demonstrated the unique cardiac transcriptomic profile upon CHIKV infection (color scale: log2fold change). (C) Significantly up- or downregulated genes as a result of CHIKV infection in specific compartments.