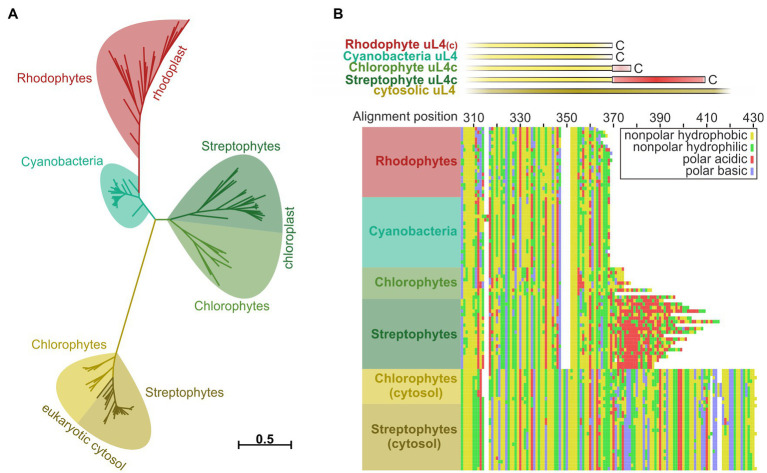
Figure 5.
Reconstruction of phylogenetic relationships and amino acid sequence comparisons between uL4 proteins of photosynthetic organisms. Phylogeny- and multiple sequence alignment-based analysis of plastidic and cytosolic uL4 sequences from chlorophytes, streptophytes, rhodophytes, and cyanobacteria. (A) Unrooted maximum likelihood tree of the homologous uL4 peptide sequence regions. Branch lengths are scaled according to the number of substitutions per site. (B) MAFFT alignment of the uL4 amino acid sequences, using the L-INS-I algorithm. Top: schematic representation of the aligned sequences. Bottom: Cropped alignment emphasizing the C-terminal regions of the bacterial/plastidic uL4/uL4c proteins. Amino acids are represented by their chemical properties: non-polar hydrophobic (yellow; A,F,I,L,M,P,V,W), non-polar hydrophilic (green; C,G,N,Q,S,T,Y), polar basic (blue; H,K,R) and polar acidic (red; D,E).