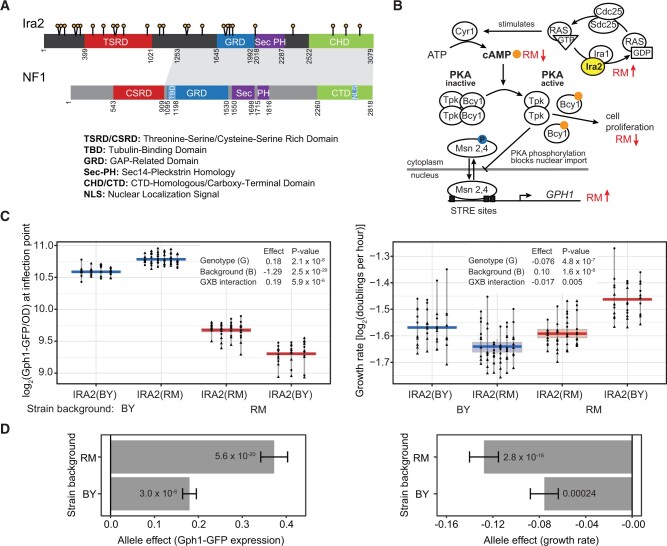
Figure 1.
RM variants within the coding region of IRA2 increase Gph1-GFP expression in trans. (A) Schematic showing nonsynonymous variants (lollipops) and domains (colored boxes) in the Ira2 protein. The NF1 schematic is modified from Bergoug et al. (2020). Amino acid positions of domain and conservation boundaries are indicated. The shaded region connects the extended regions of amino acid conservation between Ira2 (positions 1253–3079) and NF1 (positions 909–2818). (B) The role of IRA2 in the cAMP-PKA pathway including GPH1. The anticipated change in the levels or activity in the presence of the RM vs the BY allele is indicated by red arrows. Activation of PKA occurs upon binding of cAMP (orange circle) to the regulator Bcy1, resulting in its dissociation from the catalytic subunits (Tpk1, 2 or 3). See main text for more details. Figure is adapted from Santangelo (2006). (C) Effect of the IRA2 RM coding variants on Gph1-GFP expression and growth rate in the BY and RM backgrounds. For each strain, three to six transformants were phenotyped at least 10 times during two plate reader runs. Different shapes represent different plate reader runs. Lines connect measurements of the same transformant. The boxplots show the median as thicker central lines and the first and third quartiles computed on averaged phenotypes per transformant. (D) The IRA2 RM allele effect in the BY and RM backgrounds. The allele effect is the log2 of the average Gph1-GFP expression or growth rate in the presence of the RM allele minus that in the presence of the BY allele. Error bars are the standard error of the estimate. P-values are for a comparison between the RM allele and the BY allele in the designated background.