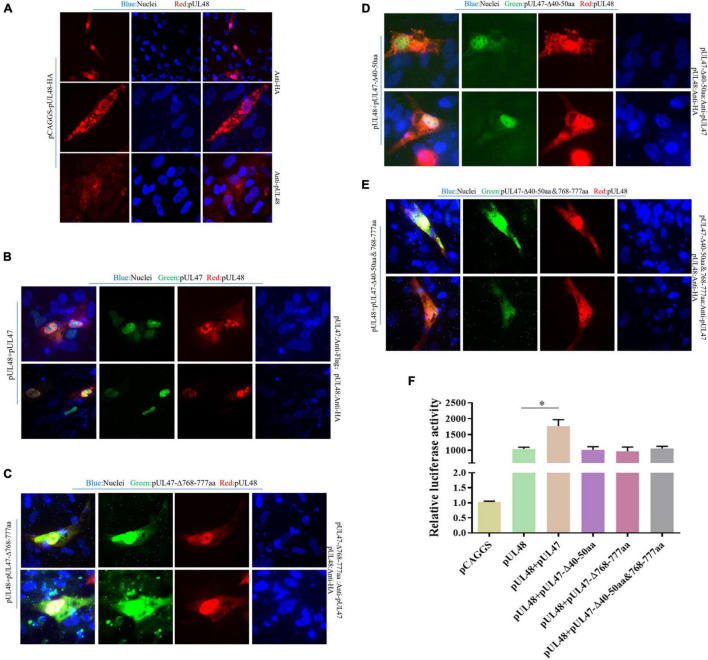
FIGURE 8.
Effects of pUL47 NLS on transcriptional activation of pUL48. (A) Cell localization of pUL48. (B) The effect of pUL47 on the nucleation of pUL48. (C) The influence of pUL47-Δ40–50 aa on pUL48 localization. (D) The influence of pUL47-Δ768–777 aa on pUL48 localization. (E) The influence of pUL47-Δ40–50 and 768–777 aa on pUL48 localization. First, pCAGGS-pUL48-HA was transfected separately. Then pCAGGS-pUL48-HA, pCAGGS-pUL47-Flag, pcDNA3.1-pUL47-Δ40–50 aa, pcDNA3.1-pUL47-Δ768–777 aa, and pcDNA3.1-pUL47-Δ40–50 and 768–777 aa with NLS deletion plasmids were cotransfected according to corresponding experimental groups, and indirect immunofluorescence assay was performed. (F) Effect of pUL47 NLS deletion on ICP4-Pro (1,235 bp) activation of pUL48. pCAGGS, pCAGGS-pUL48-HA, pCAGGS-UL47-Flag, pcDNA3.1-pUL47-Δ40–50 aa, pcDNA3.1-pUL47-Δ768–777 aa, and pcDNA3.1-pUL47-Δ40–50 and 768–777 aa were cotransfected into DEF with pGL4-ICP4-Pro (1,235 bp)-Luc and pRL-TK, except the reference plasmid pRL-TK. The transfection ratio of other plasmids with pGL4-ICP4-Pro (1,235 bp)-Luc was 1:1, and the transfection quantity of pRL-TK was 1/20 of pGL4-ICP4-Pro (1,235 bp)-Luc. Cell samples were collected 24 h after transfection, and ICP4-Pro (1,235 bp) promoter activity was detected by a dual-luciferase reporting system. Student’s t-test was used to analyze the differences of the two groups; *p < 0.05.