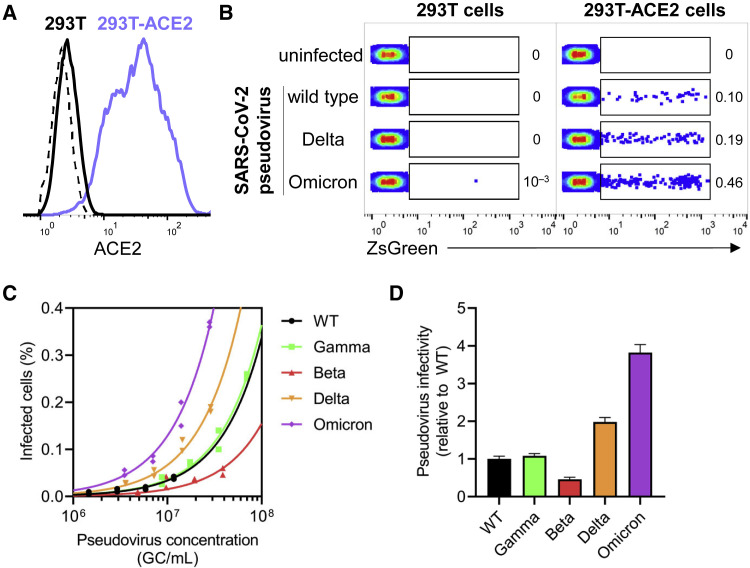
Figure 4.
SARS-CoV-2 Omicron pseudovirus demonstrates a substantial increase in infectivity of ACE2+ cells relative to other SARS-CoV-2 variants in vitro
(A) Flow cytometry histogram depicting ACE2 surface staining on parental 293T cell line (in black), and 293T-ACE2 cell line stably expressing human ACE2 (in lilac). Dashed line indicates unstained control.
(B) Representative dot plots show percentage of 293T (left panel) and 293T-ACE2 (right panel) cells that were infected with wild-type, Delta, or Omicron SaRS-CoV-2 pseudoviruses (within ZsGreen+ gate) after 48 h of co-culture. Pseudoviruses were produced in parallel and under identical conditions.
(C) Titering of SARS-CoV-2 pseudoviruses of wild-type, Gamma, Beta, Delta, and Omicron variants was performed on 293T-ACE2 cells and correlated to pseudovirus concentration in genome copies (GC)/mL as determined by qPCR. Two technical replicates (n = 2) and a linear regression were performed to fit data, given that a linear relationship of virus concentration versus infected cells can be assumed at infection rates <10%.
(D) Pseudovirus infectivity relative to wild type was measured for each SARS-CoV-2 variant in (C) by calculating fold change in slope from (C) for each pseudovirus relative to wild type. Bars and error bars depict mean and standard error of the mean.