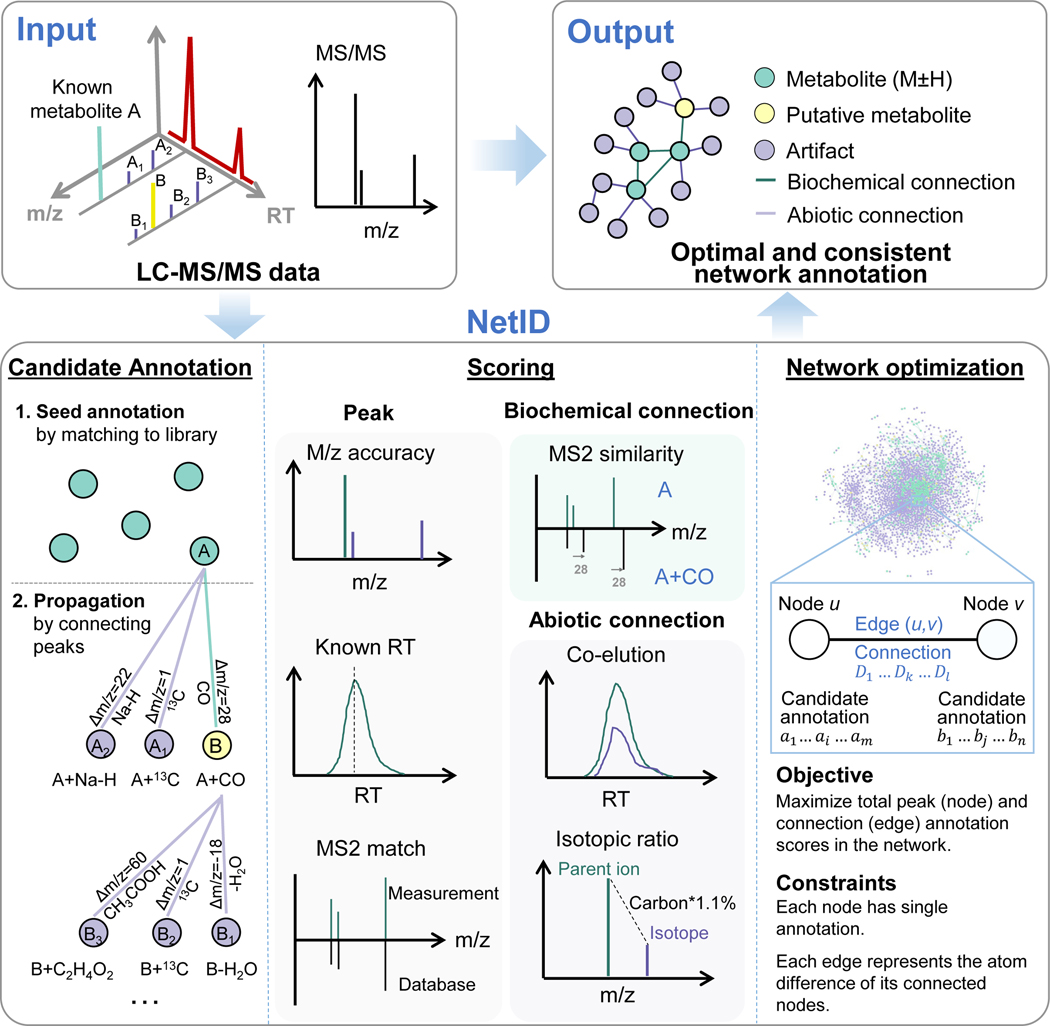
Figure 1. A global network optimization approach for untargeted metabolomics data annotation (NetID).
The input data are LC-MS peaks with m/z, retention times, intensities and optional MS2 spectra. The output is a molecular network with peaks (nodes) assigned with unique formulae and connected by edges reflecting atom differences arising either through metabolism (biochemical connection) or mass spectrometry phenomenon (abiotic connection). Peaks are classified as “metabolite” (M+H or M-H peak of formula found in selected metabolomics database, e.g. HMDB), “putative metabolite” (formula not found in database but with biochemical connection to a metabolite), or “artifact” (only abiotic connection to a metabolite). NetID algorithm involves three steps. Candidate annotation first matches peaks to database formulae. These seed annotations are then extended through edges to cover most nodes, with the majority of nodes receiving multiple formula annotations. Each node and edge annotation are then scored based on match to known masses, retention times, and MS/MS fragmentation patterns. Global network optimization maximizes sum of node scores and edge scores, while enforcing a unique formula for each node and a unique transformation relationship for each edge.