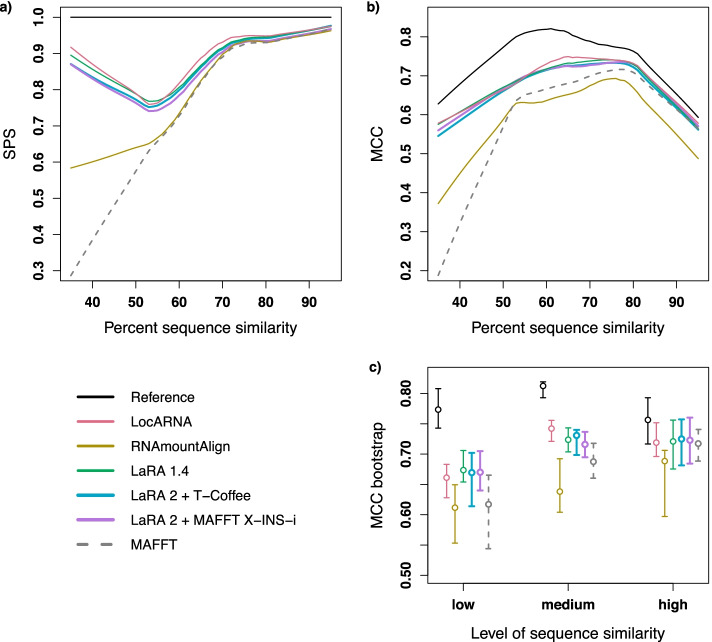
Fig. 4.
a Sum-of-pairs score and b Matthews correlation coefficient are shown for different tools dependent on the sequence similarity. The tools were run on 388 alignments of the BRAliBase 2.1 data set (without SRP) and the curves were generated with a lowess function on the results. In order to show the MCC performance of MAFFT as a sequence alignment tool, as well as of the reference alignment from BRAliBase, we calculated the best secondary structure of the alignments with RNAalifold [35]. c bootstrap percentile confidence intervals and medians for the MCC values. The first axis represents the sequence similarity in three groups: low (), medium ( and ) and high (), as annotated in BRAliBase [50]. For each group we bootstrapped 1000 samples of the MCC experiment