TABLE 2.
Microglia phenotypes in patients with Alzheimer’s disease revealed by single-cell technologies.
| References | Patients | Method | Brain region | Microglia state/cluster (gene signature) |
| Del-Aguila et al., 2019 | 3 patients with Mendelian or sporadic AD | snRNA-seq | parietal lobes |

|
| Mathys et al., 2019 | 48 AD patients | sRNA-seq | Prefrontal cortex |
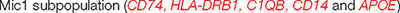
|
| Grubman et al., 2019 | 6 AD patients | sRNA-seq | Entorhinal cortex |

|
| Zhou et al., 2020 | 11 AD patients with TREM2-CV and 10 bearing TREM2-R62H | snRNA-seq | Dorsolateral prefrontal cortex |

|
| Lau et al., 2020 | 12 AD patients | snRNA-seq | Prefrontal cortex | Reduced microglia subpopulation that expresses genes which participate in synaptic pruning (C1QA, C1QB and C1QC, encoding complement component 1q) or encode cytokine receptors (IL4R and IL1RAP) |
| Nguyen et al., 2020 | 15 AD patients | snRNA-seq | Dorsolateral prefrontal cortex |
4 microglia subpopulations: homeostatic, motile, amyloid responsive, and dystrophic microglia |
| Gerrits et al., 2021 | 10 AD donors with only Aβ pathology or both Aβ and tau pathology |
snRNA-seq | Occipital or occipitotemporal cortex |
AD1 microglia population express DAM genes, and are enriched with AD risk genes and correlated with tissue Aβ load; AD2 microglia express glutamate receptor GRID2 and are correlated with tau pathology |
Blue, downregulated signature genes; red, upregulated signature genes. AD, Alzheimer’s disease; DAM, disease-associated microglia; Aβ, β-amyloid.
