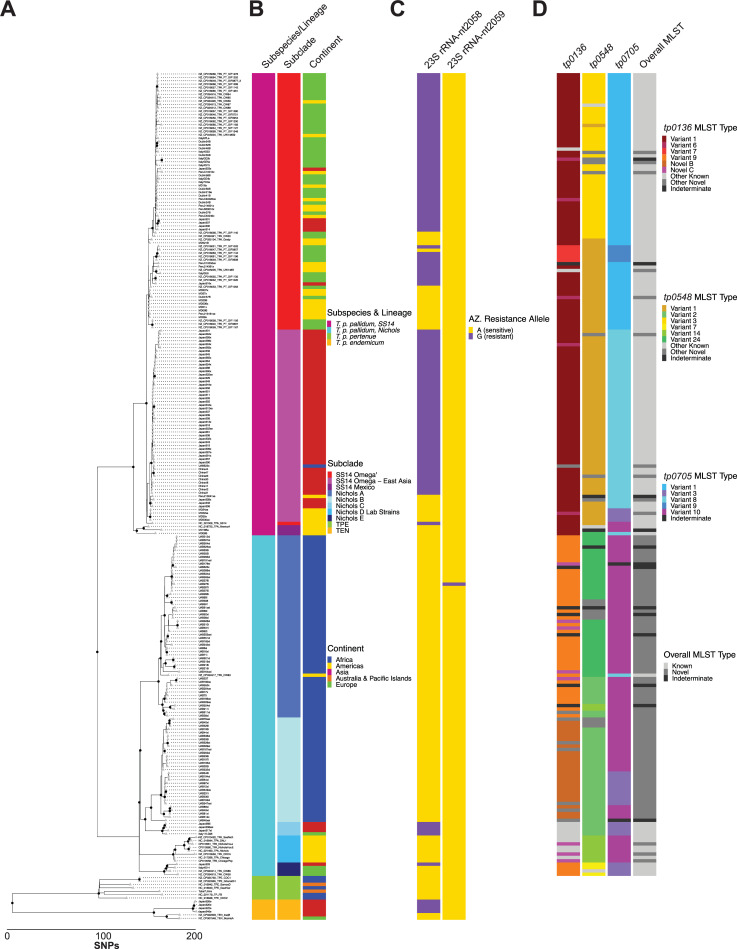
Fig 1. Recombination-masked whole genome phylogeny of T. pallidum patient isolates.
A) Whole genomes were MAFFT-aligned, recombination-masked, and maximum-likelihood phylogeny determined. Tips are shown as grey triangles and nodes with >0.95 support from 1000 ultrafast bootstraps shown as black circles. B) Subspecies/lineage, subclade, and continent of origin of all samples included in phylogeny. C) Azithromycin sensitivity/resistance as conferred by the 23S rRNA 2058/2059 alleles. Data represents alleles at both rRNA loci. D) MLST subtypes, including novel sequences, for tp0136, tp0548, and tp0705, as well as whether the three alleles constitute a known or novel MLST. Top 6 most abundant sequences at each locus are colored, while other less abundant known and novel sequences are grouped and colored in light and medium grey, respectively. Sequences containing N bases are denoted as indeterminate and shown in dark grey. Expanded metadata for all samples is included in S1 Data.