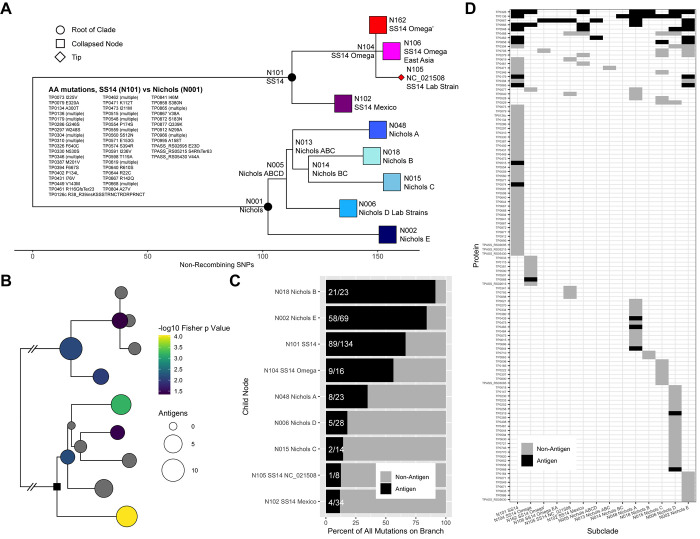
Fig 4. Coding mutations in the T. pallidum subsp. pallidum phylogeny.
A) Whole genome ML phylogeny of TPA, with tips collapsed to the subclade node. Open reading frames of inferred ancestral sequences for each node were annotated based on the SS14 reference sequence NC_021508. Coding mutations, including for putative recombinant genes, for each child node were determined relative to its parent node (complete list in S4 Data). Loci with amino acid differences (n = 49 loci, n = 134 individual AA mutation events) in the SS14 ancestral clade node (N101) are shown relative to the Nichols ancestral node (N001). B) Positions are equivalent to those shown in A. Black square represents the Nichols Ancestral Node (N001). Number of antigens with coding mutations on each child node relative to parent node. Color represents p value of for overrepresentation by Fisher’s Exact test of antigens among all mutated proteins per branch; those in grey have a p value > 0.05. C) Percentage of total individual mutation events per branch. Raw numbers of mutation events in antigens per total mutation events are shown for each branch. D) Tile plot showing mutated proteins in the ancestral node for each subclade relative to its parent node, colored by antigen or not. Proteins are arranged by number of subclades bearing mutations. Data is recapitulated in S3 Data.