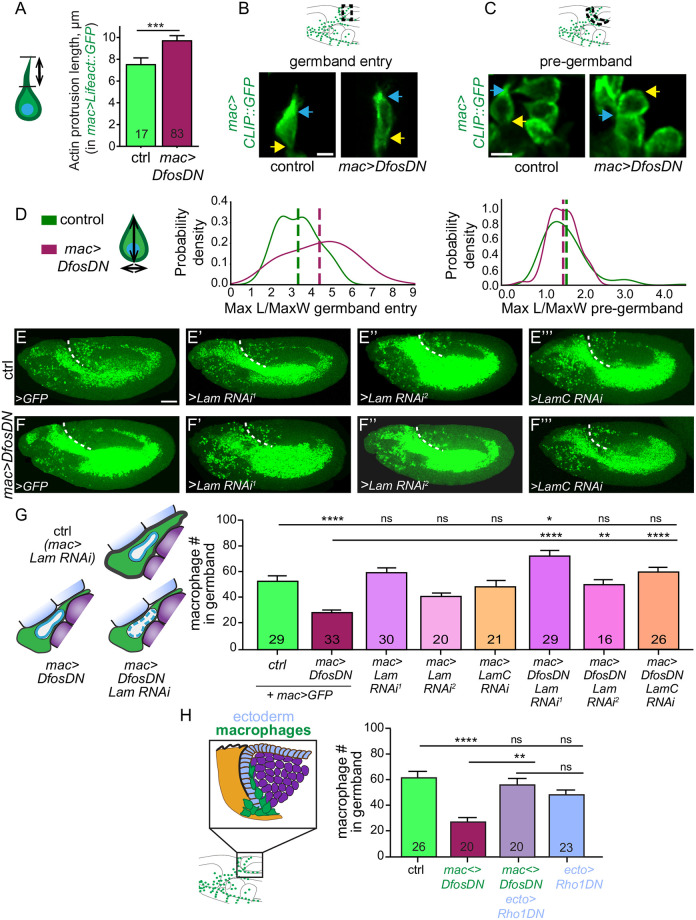
Fig 5. Dfos aids macrophage gb invasion against the resistance of surrounding tissues and buffers the nucleus.
(A) Quantification from live embryos shows that the length of the F-actin protrusion of the first entering macrophage is longer in macrophages expressing DfosDN. p = 0.011. The F-actin protrusion labeled with srpHemo-Gal4 driving UAS-LifeAct::GFP was measured in the direction of forward migration (see schematic). SD: 2.4, 3.7. (B-C) Stills from 2-photon movies of St 11 embryos showing (B) the first macrophages entering the gb and (C) macrophages in the pre-gb zone in the control and in a line expressing DfosDN in macrophages. Microtubules are labeled with srpHemo-Gal4 driving UAS-CLIP::GFP. A blue arrow indicates the front and a yellow arrow indicates the rear of the macrophage. Schematics above indicate where images were acquired. (D) Schematic at left shows macrophage measurements: vertical line for the maximum length and horizontal line for the maximum width. Histograms show the probability density distributions of the aspect ratios (maximum length over maximum width) of the first macrophage entering the gb (left) and macrophages in the pre-gb (right). Macrophages expressing DfosDN are more elongated than the controls. Control vs. mac>DfosDN aspect ratios at gb entry p = 0.0011, in pre-gb p = 0.53. SD: in gb 1.0, 1.6; in pre-gb 0.5, 0.5. (E-F”’) Confocal images of St 12 embryos expressing RNAi against Lamin or LaminC in macrophages in (E-E”’) the control, or (F-F”’) in embryos also expressing DfosDN in macrophages. srpHemo-GAL4 used as driver. Lam RNAi1: GD45636. Lam RNAi2: KK107419. LamC RNAi: TRiP JF01406. (G) Macrophage RNAi knockdown of Lamins, which can increase nuclear deformability did not affect macrophages numbers in the gb in the control. In embryos in which macrophages expressed DfosDN, Lamin knockdown rescued their reduced numbers in the gb. Control vs. DfosDN p < 0.0001. Control vs. Lam RNAi1 p > 0.99, vs. Lam RNAi2 p = 0.83, vs. LamC RNAi p > 0.99. Control vs. DfosDN, Lam RNAi1 p = 0.024, vs. DfosDN, Lam RNAi2 p > 0.99, vs. DfosDN, LamC RNAi p > 0.99. DfosDN vs. DfosDN, Lam RNAi1 p < 0.0001, vs. DfosDN, Lam RNAi2 p = 0.0049, vs. DfosDN, LamC RNAi p < 0.0001. SD: 22, 10, 19, 11, 21, 23, 16, 20. (H) Expressing DfosDN in macrophages reduces their number in the gb. Concomitantly reducing tissue tension in the ectoderm (light blue in schematic) through Rho1DN substantially rescues invasion. srpHemo-QF QUAS control (mac<>) governed macrophage expression and e22c-GAL4 ectodermal (ecto>). Control vs. mac<>DfosDN p < 0.0001 (56% reduction), vs. mac<>DfosDN; ecto>Rho1DN p > 0.99, vs. ecto>Rho1DN p = 0.11. mac<>DfosDN vs. mac<>DfosDN; ecto>Rho1DN p < 0.0001, vs. ecto>Rho1DN p = 0.0044. mac<>DfosDN; ecto>Rho1DN vs. ecto>Rho1DN p > 0.99. SD: 23, 16, 21, 18. Macrophages are labeled in B-C by srpHemo-Gal4 driving UAS-CLIP::GFP, and in E-F’” by srpHemo-Gal4 UAS-mCherry::nls. ****p < 0.0001, ***p < 0.005, **p < 0.01, *p < 0.05. Unpaired t test was used for A, one-way ANOVA with Tukey post hoc for G-H. The number shown within the column corresponds to measurements in A, and analyzed embryos in G-H. Scale bar 5 μm in B-C, and 50 μm in E-F”’. The data underlying the graphs can be found in S1 Data. ctrl, control; gb, germband; ns, not significant; RNAi, RNA interference.