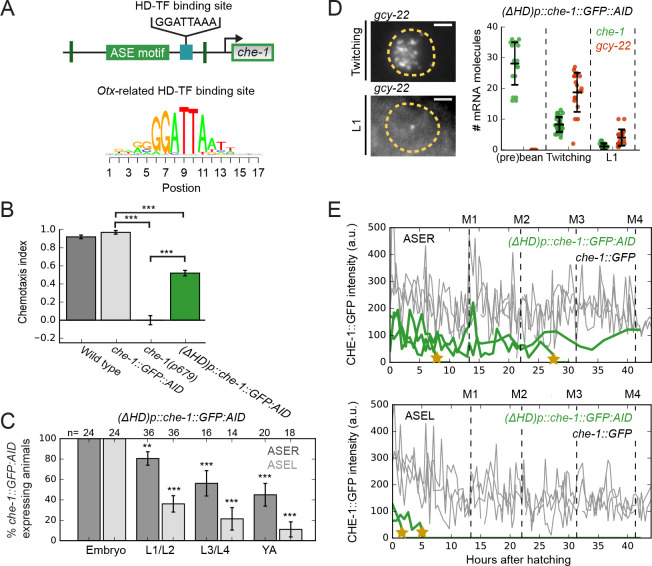
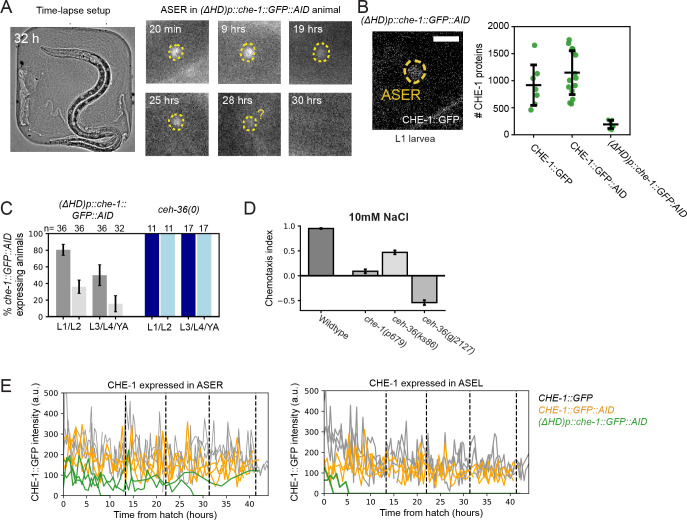
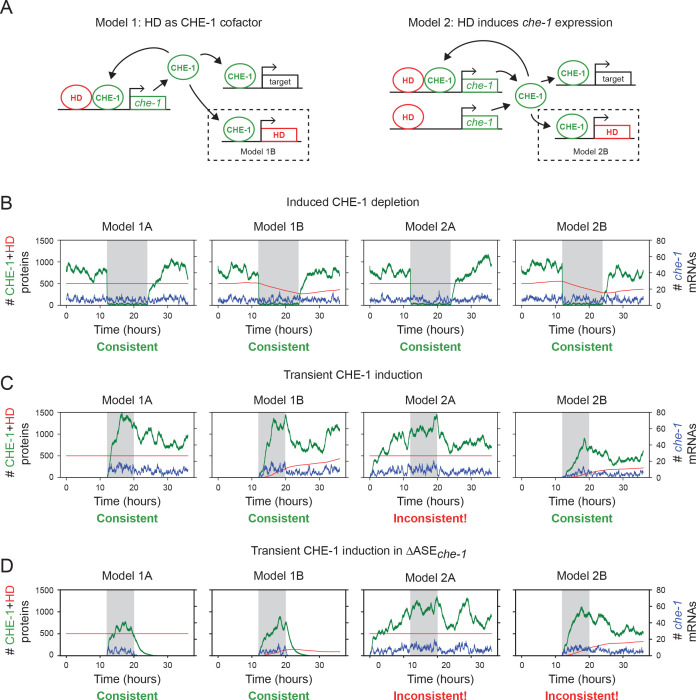
Figure 7. An Otx-related homeodomain transcription factor binding site involved in long-term maintenance of ASE cell fate.
(A) Position of an Otx-related homeodomain transcription factor (HD-TF) binding site in the che-1 promoter. Green lines indicate the positions of the 59 bp flanks surrounding the ASE motif. HD-TF binding site depicted as a sequence logo. In (ΔHD)p::che-1::GFP::AID animals, the HD-TF-binding site is deleted in the che-1::GFP::AID background. (B) Average chemotaxis index for response to 10 mM NaCl, of wild-type, che-1::GFP::AID and che-1(p679) animals, and (ΔHD)p::che-1::GFP::AID mutant. Deleting the HD-TF-binding site caused a decreased response to NaCl. (C) Fraction of (ΔHD)p::che-1::GFP::AID animals expressing CHE-1::GFP::AID in ASER (dark gray) and ASEL (light gray) at different developmental stages. CHE-1::GFP::AID is progressively lost during development. (D) che-1::GFP::AID and gcy-22 mRNA levels in (ΔHD)p::che-1::GFP::AID animals quantified by smFISH. Expression was similar to wild-type until late-stage, twitching embryos, but fell rapidly in newly-hatched L1 larvae. Scale bar: 2 μm. (E) CHE-1::GFP expression dynamics in single che-1::GFP (grey) and (ΔHD)p::che-1::GFP::AID (green) animals during larval development. Approximate timing of molts is indicated M1-M4. CHE-1::GFP::AID expression in (ΔHD)p::che-1::GFP::AID animals was lost in a rapid and stochastic manner, at different times during development. Error bars represent S.E.M (B,D) or S.D. (C). n ≥ 10. **p < 0.01, ***p < 0.001.