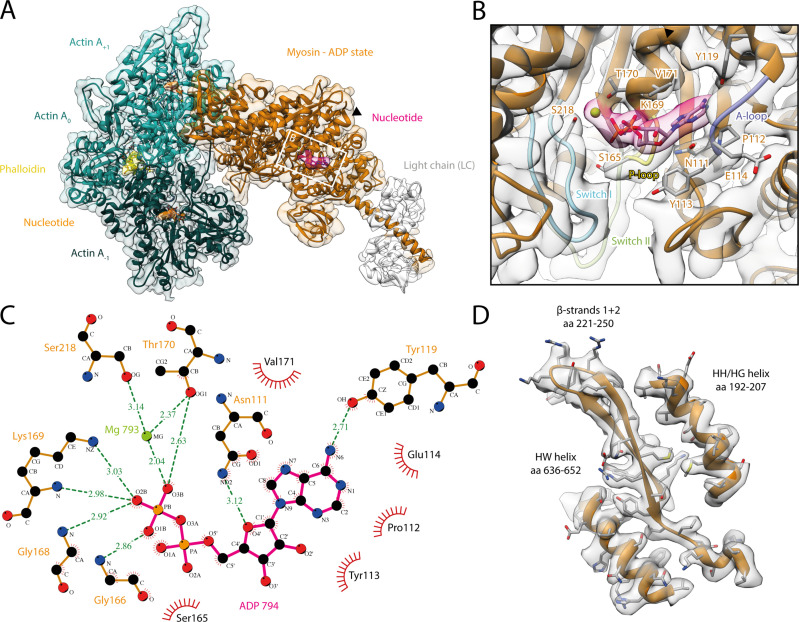
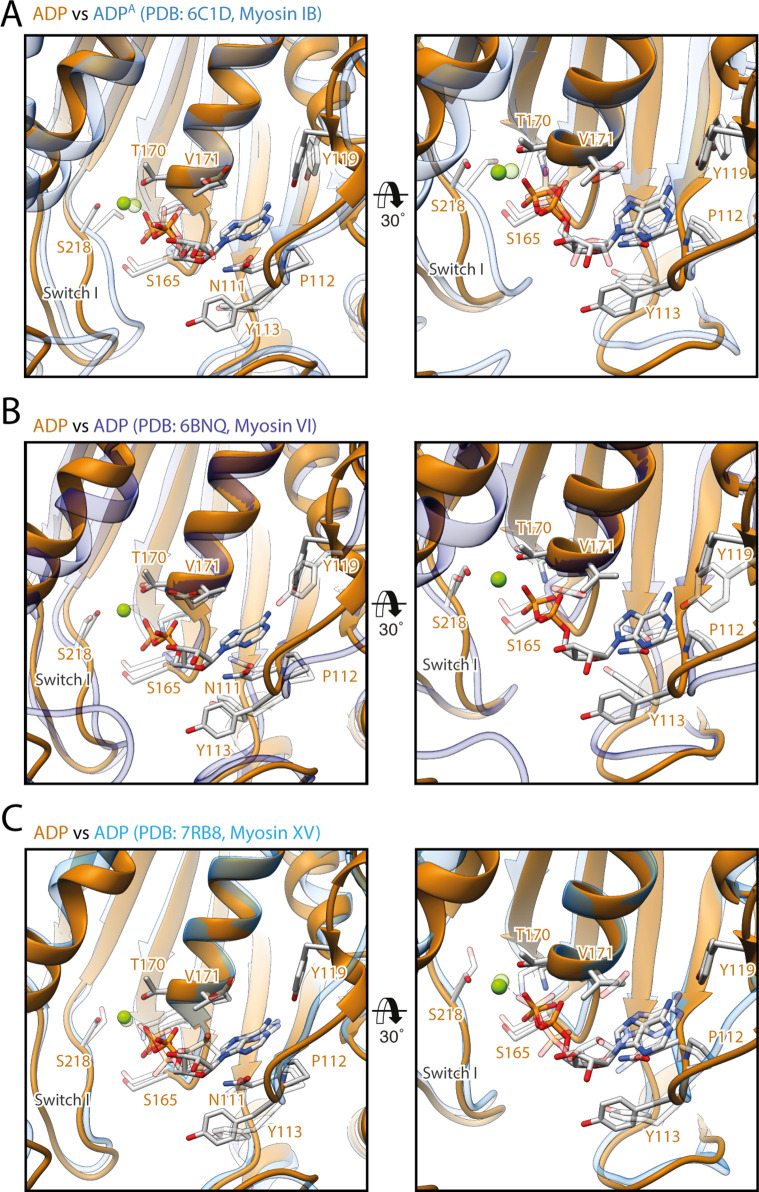
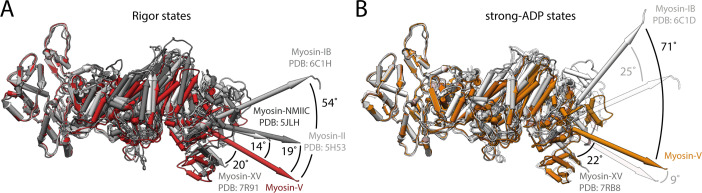
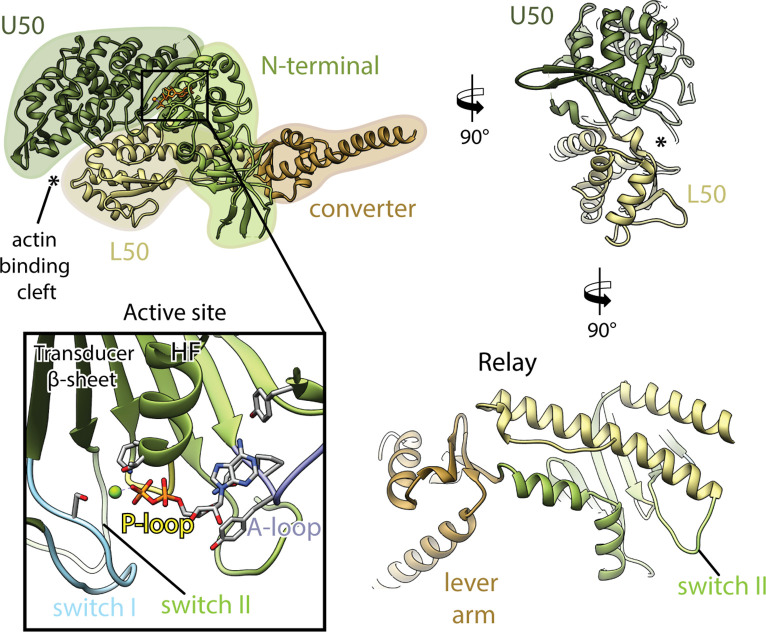
Figure 1. Structure and active site of the aged actomyosin-V complex bound to ADP.
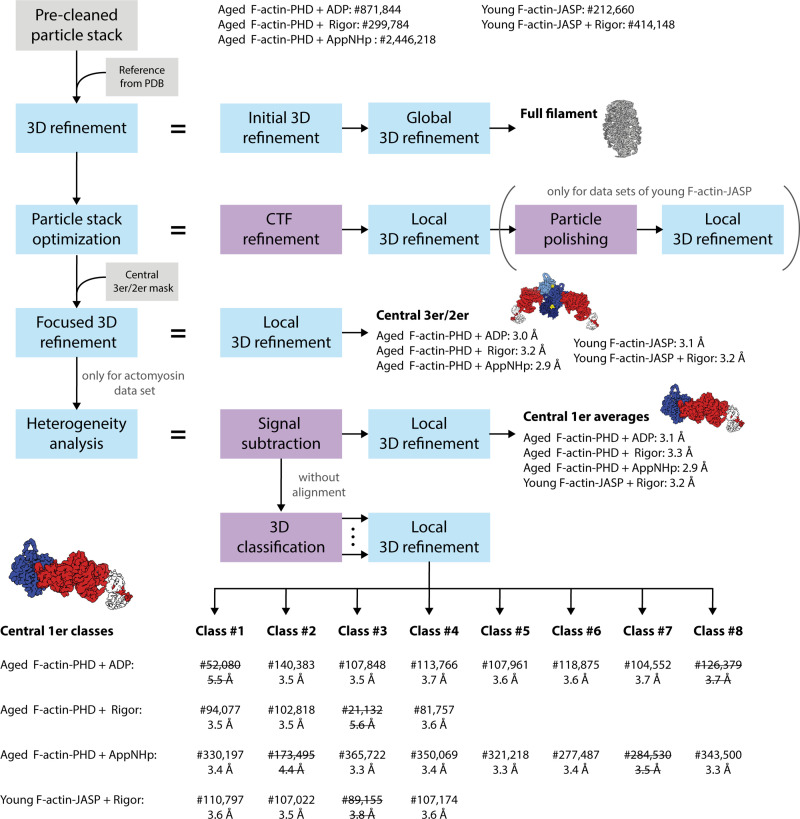
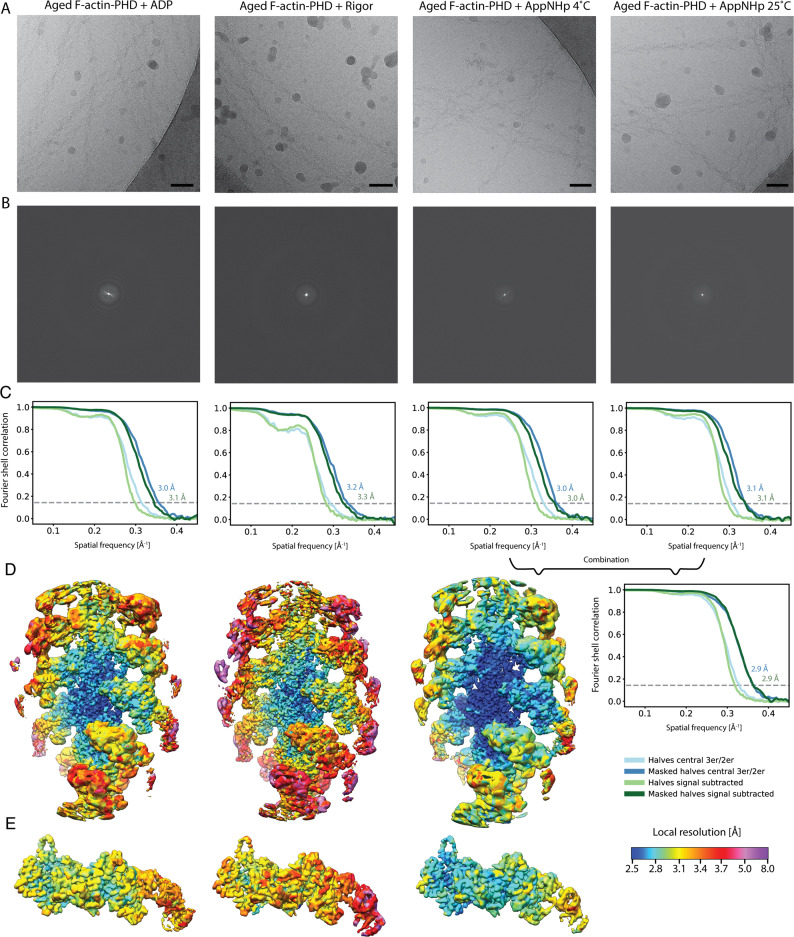
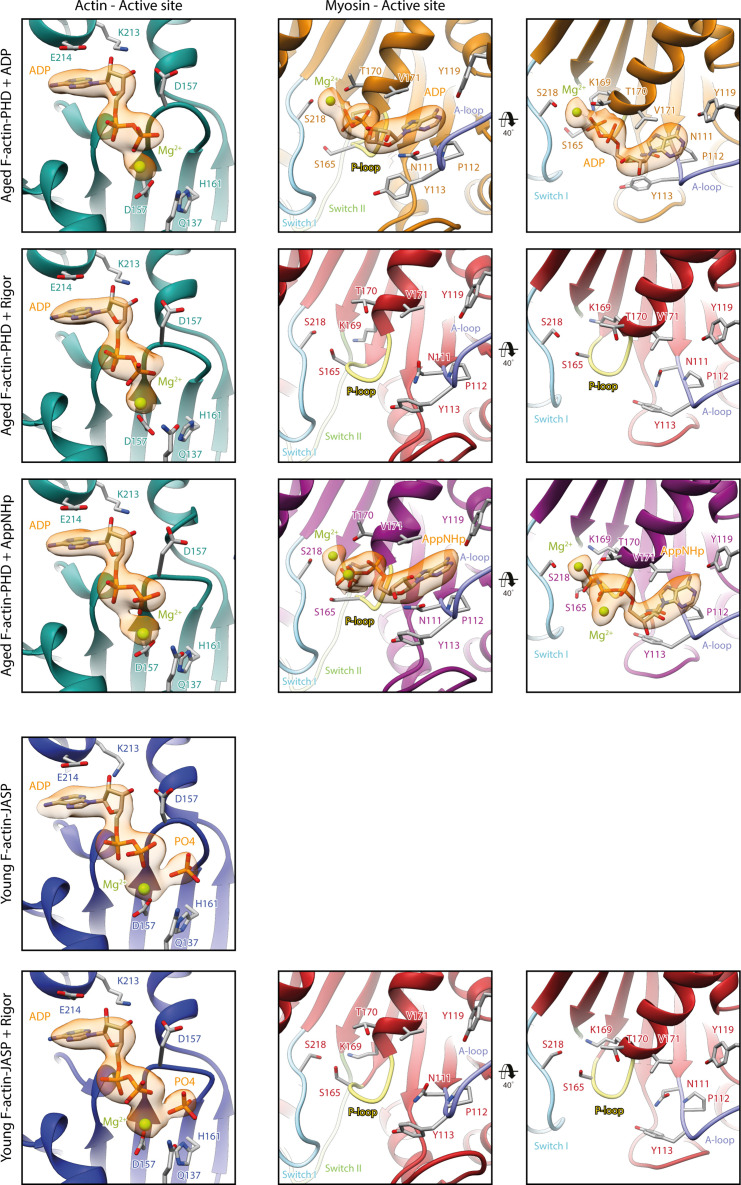
(A) Atomic model and LAFTER density map of the central myosin-V-LC subunit (orange, LC: white) bound to aged F-actin-PHD (shades of sea green, three subunits shown, A-1 to A+1). Nucleotides and PHD are highlighted in orange, pink, and yellow, respectively. The HF helix is marked by a black arrowhead. (B) Close-up view of the myosin active site consisting of the P-loop (yellow, 164–168), switch I (blue, aa 208–220), switch II (green, aa 439–448), and the A-loop (purple, aa 111–116). Only side chains involved in the binding of ADP are displayed, also see Figure 1—figure supplement 3. (C) 2D protein-ligand interaction diagram illustrating the coordination of Mg2+-ADP by hydrogen bonds (dashed green lines) and hydrophobic interactions (red rays). (D) Illustration of the model-map agreement within a central section of myosin. Most side chains are resolved by the post-refined density map (transparent gray). See Figure 1—video 1 for a three-dimensional visualization and Figure 1—figure supplements 1–2 for an overview of the processing pipeline and the cryo-EM data, respectively. A comparison of the strong-ADP state of different myosins can be found in Figure 1—figure supplements 4 and 5. Figure 1—figure supplement 6 illustrates the domain architecture of myosin.