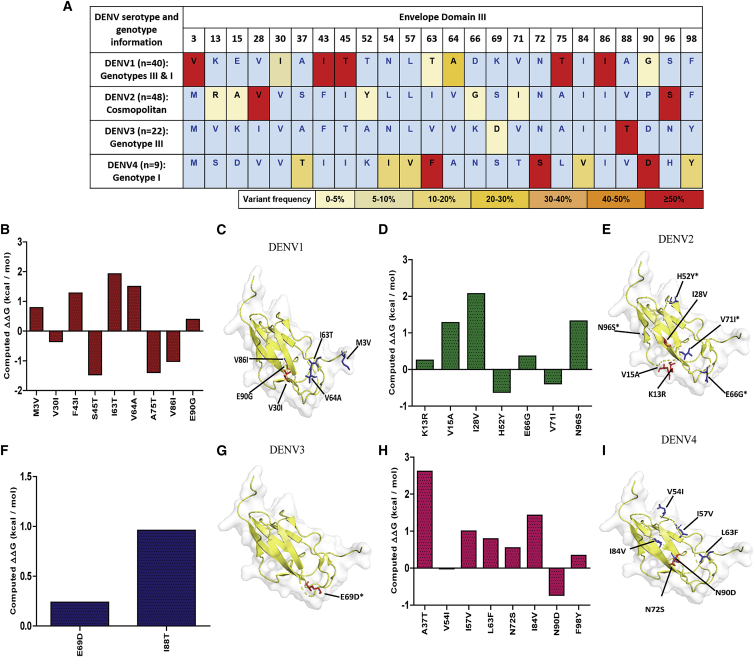
Figure 2.
DENV EDIII amino acid variation in our study clinical isolates for all four serotypes
(A) Frequency-based representation of all the variable sites for each serotype within our study sequences. Amino acid variant residues identified relative to NC_001477 (DENV1), NC_001474 (DENV2), NC_001475 (DENV3), and NC_002640 (DENV4). Colors were assigned based on the percentage of the frequency of mutations in the given clinical isolates. (B, D, F, and H) FoldX stability calculations for mutations in DENV1, DENV2, DENV3, and DENV4 strains, respectively. (C, E, G, and I) Amino acid variants on B cell and T cell epitopes are spotted on EDIII PDB structure (ribbon). The stick and ball representation on the PDB structure (PDB: 3IRC) indicates a mutation at the particular position. The presence of mutations in the B cell or T cell epitopes is shown by arrows. B cell mutations are shown in red, and T cell mutations are shown in blue. Asterisks (∗) signify mutations in both B and T cell epitopes.