Abstract
Niemann-Pick disease type C (NP-C) (OMIM#257220) is a rare lysosomal storage disorder caused by pathogenic variants in either the NPC1 or NPC2 genes. It manifests with a wide spectrum of clinical symptoms and variable age of onset. We studied the impact of the frequent polymorphic variant c.2793 C > T (p.Asn931 = ), located in the donor splice site (SS) of NPC1 exon 18 on the penetrance of the rare synonymous variant c.2727 C > T (p.Cys909 = ), identified in two 55 y.o. twins with an adult onset form of NP-C. The patients’ diagnosis was supported by biochemical analysis and positive filipin test. Analysis of the patients’ cDNA showed that the c.2727 C > T variant leads to cryptic donor SS activation and frameshift deletion in the NPC1 exon 18. However, the minigene assay demonstrated that this exon shortening takes place only in the presence of the frequent polymorphic variant c.2793 C > T. Results of the transcript specific qPCR showed that only the presence in the NPC1 exon 18 of both variants leads to significant decrease of wild type (WT) transcript isoform.
Subject terms: Neurological disorders, RNA splicing
Introduction
NP-C is an autosomal recessive disorder caused by pathogenic variants in either the NPC1 or NPC2 genes in ~95% and 5% cases respectively [1]. NPC1 encodes a transmembrane protein involved in transport of lipids, particularly cholesterol, from late endosomes or lysosomes [2]. The loss of its function leads to excessive accumulation storage of unesterified cholesterol, sphingomyelin, phospholipids and glycolipids in liver, spleen, and central nervous system. Subsequent neuropathological defects include Alzheimer’s-like neurofibrillary tangles, neuronal degeneration, neuroaxonal dystrophy, and demyelination with white matter tracts severely affected, especially in corpus callosum.
NP-C has a broad range of clinical symptoms and severity, from progressive fatal neonatal disorder to milder adulthood and later onset forms. Progressive neurodegeneration is a highlight of the disease and determines its severity. Neurological symptoms include motor development delay in early childhood; gaits, falls, clumsiness, cataplexy, and school problems in later childhood and adolescence; psychiatric illness and dementia in adult variants. Due to the predominating neuropsychiatric and cognitive symptoms, adult forms of NP-C are probably highly underestimated and patients are often misdiagnosed [3, 4]. According to recent publications, the prevalence of adult- and adolescent-onset forms of NP-C could be much higher than 1:100000 for the “classical” incidence with early severe phenotype [1, 5, 6].
In this article we present the detailed clinical description, biochemical data and functional analysis of the novel complex allele (CA) c.[2727 C > T;2793 C > T] in the NPC1 gene of two dizygotic twins with an adult-onset form of NP-C.
Materials and methods
All variants are called according to NPC1 canonical transcript NM_000271.5 and the Human Genome Variation Society nomenclature. Exons are numbered according to reference sequence NG_012795.1.
Description of methods, patients clinical and biochemical data are available in Supplementary file 1.
Results
P1 and P2 (55 y.o. twins) were suspected for NP-C disease based on a characteristic set of clinical symptoms presented in Supplementary Table 1. Elevation of the metabolites Lysosphingomyelin-509, cholestan-3β, 5α, 6β-triol and 7-ketocholesterol were detected and subsequent genetic testing of the NPC1 and NPC2 genes was performed.
The next-generation sequencing of NPC1 and NPC2 exons revealed two compound heterozygous variants in NPC1: complete loss-of-function variant c.2196dup (p.Ala732fs*30), described previously [7], and rare synonymous variant c.2727 C > T (p.Cys909 = ) with very low allele frequency in gnomAD (1.06e−5). The validation of these variants by Sanger sequencing also revealed the presence of c.2793 C > T (p.Asn931 = ) polymorphism in homozygous state (gnomAD allele frequency 4.94e−1), located in the NPC1 exon 18 donor SS. The DNA analysis of patient’s kindreds demonstrated that they are heterozygous carriers either of c.2727 C > T or c.2196dup variants, confirming their trans-position (Fig. S1).
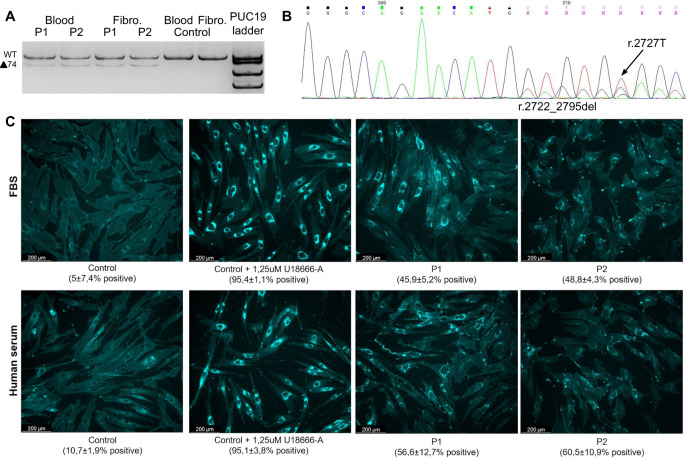
The RT-PCR performed on RNA samples from blood and fibroblasts showed that patients have an additional lower-molecular-weight band together with 552 b.p. band of WT product (Fig. 1A). Sanger sequencing revealed that this band presents a 74 b.p. deletion r.2722_2795del (p.Val908Tyrfs*32) in exon 18, caused by activation of cryptic exonic SS (Fig. 1B). Also, the presence of r.2727 T variant in chromatogram indicates that the c.2727 C > T variant is “leaky” and gives significant amount of full-length transcript isoforms. In order to confirm the NP-C diagnosis, we performed the filipin staining test. After cholesterol depletion, skin fibroblast cultures from both patients were challenged either with a medium containing 10% of FBS and 10% of LDLP-enriched human serum. Filipin staining showed increased intracellular accumulation of non-esterified cholesterol in perinuclear vesicles of both patients with a smaller number and size of fluorescent perinuclear vesicles and a lower overall level of fluorescence compared to the positive control cells, treated with U18666A, consistent with a “variant” pattern according to Vanier and Latour classification [8] (Fig. 1C).
Fig. 1. Patients’ mRNA analysis and filipin staining.
A Visualization of PCR products from amplification of patients NPC1 cDNA including exons 16–20. B The Sanger sequencing chromatogram demonstrating 74 b.p. deletion r.2722_2795del (p.Val908Tyrfs*32) caused by cryptic SS activation. The presence of r.2727T variant in chromatogram indicates that the c.2727 C > T variant is “leaky” and gives significant amount of full-length transcript isoforms. C Filipin staining of patients’ fibroblasts demonstrate the “variant” pattern.
To validate the effect of c.2727 C > T variant on splicing, and also to estimate the proportion of transcript isoforms in the absence of nonsense-mediated mRNA decay and the second allele we decided to perform minigene assay. Furthermore, since the c.2793 C > T (p.Asn931 = ) polymorphism is located in the WT donor SS of the same exon 18, we hypothesized whether this c.2793 C > T variant could affect the balance between WT and mutant SS activity or even decrease the strength of WT site below a certain threshold required for the cryptic site activation.
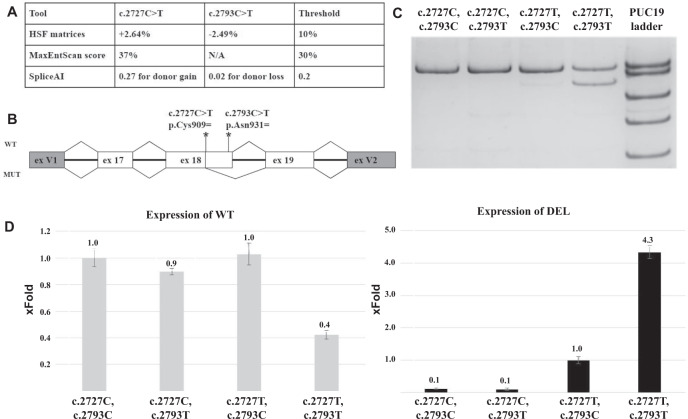
The bioinformatic analysis with various tools demonstrated no or low impact of both variants on splicing (Fig. 2A). MaxEntScan algorithm integrated in HSF wasn’t able to predict the effect of c.2793 C > T due to the very low score of the native donor splice site, although the same algorithm stand-alone predicts a dramatic reduction of its strength −0.68 > −3.03.
Fig. 2. Functional analysis of the complex allele.
A The results of bioinformatic analysis (HSF—Human Splicing Finder 3.1). B The structure of the minigene and location of variants. C The visualization of PCR products from the minigene assay. D The box plot representing the relative expression of wild type transcript isoform (WT) and the one harboring 74 b.p. deletion (DEL) in minigenes with various combinations of studied variants.
To assess the impact of these variants on normal splicing alone and their relationship with each other, we built 4 minigene constructs bearing various combinations of these variants.
17, 18, 19 exons of NPC1 gene were amplified from the patient’s DNA and cloned into pSpl3-Flu2 vector. The RT-PCR analysis of cells transfected with these minigenes demonstrated the same splicing pattern as was observed from amplification of the patients’ cDNA.
At the next step, using site directed mutagenesis, we built two additional minigenes, one of which contains c.2727 C > T but lacks c.2793 C > T and other one that lack both c.2727 C > T and c.2793 C > T. The visualization of splicing products showed that only the minigene containing both c.2727 C > T and c.2793 C > T gives a significant amount of aberrant transcripts with 74 b.p. deletion (Fig. 2C).
To make quantitative assessment, the real-time PCR technique was applied. Minigene with both variants in cis showed 58% reduction of WT isoform and the 4.33 fold more mutant isoform, compared to c.2727 C > T alone (Fig. 2D).
Discussion
The existence of CAs partially explains the phenotypic diversity for a number of genes [9–12]. In most described cases the functional effect of these variants is realized on the protein level, thus making it difficult to clarify their interactions and role in pathogenesis. Furthermore, the detection of CA itself is hampered by the limitations of common genetic screening methods e.g. filtering of frequent variants in the process of analyzing NGS data.
The CAs which alters splicing are extremely underrepresented in literature, although their effect could be predicted bioinformatically and their functional analysis is much easier compared to those which change the protein function. Furthermore, CAs affecting splicing should be located relatively close to each other on DNA sequence, making it easier for a researcher to suspect its interaction. Deep intronic variants in cis could strengthen the cryptic SSs or create the canonical dinucleotides, leading, for example, to pseudoexon inclusion [13]. Frequent or rare variants in the native SSs could change its strength and modify the effect of those variants, which activate the cryptic SSs or alters the motifs of splicing enhancers and promotes exon skipping.
Our results provide strong evidence that the frequent polymorphic variant c.2793 C > T, located in the donor SS of NPC1 exon 18 significantly affects the activity of cryptic SS created by c.2727 C > T variant. This data is supported by the bioinformatic analysis, which demonstrated low or no impact of this variant on splicing alone. The significant amount of normally spliced mRNA, observed in the minigene assay, in the presence of typical loss-of-function variant on the second allele correlates well with patient’s adult onset phenotype. Also, the microscopy examination of filipin staining test demonstrated the “variant” pattern, which is common for adult onset form of NP-C.
Overall, we identified the novel rare CA c.[2727 C > T;2793 C > T] in NPC1, characterized its deleterious effect on mRNA level and the role in NP-C pathogenesis. We highlight the necessity of analyzing the raw sequence data in cases where the identified variant leads to the cryptic SS activation, as any additional genetic variants in close proximity could significantly affect the ratio of transcript isoforms and lead to a misconception in genotype–phenotype correlations or even a wrong diagnosis.
Supplementary information
Acknowledgements
The data that support the findings of this study is submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) (Accession number—VCV000867233.3). A part of the research was done using equipment of Core Facility of Koltzov Institute of Developmental Biology RAS.
Funding
The research was carried out within the state assignment of the Ministry of Science and Higher Education of the Russian Federation for RCMG.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Ethics
The study was approved by the local ethics committee of the Federal State Budgetary Institution “Research Centre for Medical Genetics” (the approval number 2015-5/3).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-021-00898-7.
References
- 1.Patterson MC, Hendriksz CJ, Walterfang M, Sedel F, Vanier MT, Wijburg F, et al. Recommendations for the diagnosis and management of Niemann-Pick disease type C: an update. Mol Genet Metab. 2012;106:330–44. doi: 10.1016/j.ymgme.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Vance JE. Lipid imbalance in the neurological disorder, Niemann-Pick C disease. FEBS Lett. 2006;580:5518–24. doi: 10.1016/j.febslet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Burlina A. Niemann-Pick disease type C: introduction and main clinical features. J Neurol. 2014;261:S525–7. doi: 10.1007/s00415-014-7382-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geberhiwot T, Moro A, Dardis A, Ramaswami U, Sirrs S, Marfa MP, et al. Consensus clinical management guidelines for Niemann-Pick disease type C. Orphanet J Rare Dis. 2018;13:50. doi: 10.1186/s13023-018-0785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jahnova H, Dvorakova L, Vlaskova H, Hulkova H, Poupetova H, Hrebicek M, et al. Observational, retrospective study of a large cohort of patients with Niemann-Pick disease type C in the Czech Republic: a surprisingly stable diagnostic rate spanning almost 40 years. Orphanet J Rare Dis. 2014;9:140. doi: 10.1186/s13023-014-0140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wassif CA, Cross JL, Iben J, Sanchez-Pulido L, Cougnoux A, Platt FM, et al. High incidence of unrecognized visceral/neurological late-onset Niemann-Pick disease, type C1, predicted by analysis of massively parallel sequencing data sets. Genet Med. 2016;18:41–8. doi: 10.1038/gim.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stampfer M, Theiss S, Amraoui Y, Jiang X, Keller S, Ory DS, et al. Niemann-Pick disease type C clinical database: cognitive and coordination deficits are early disease indicators. Orphanet J Rare Dis. 2013;8:35. doi: 10.1186/1750-1172-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vanier MT, Latour P. Laboratory diagnosis of Niemann–Pick disease type C: The filipin staining test. Methods Cell Biol. 2015;126:357–75. doi: 10.1016/bs.mcb.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 9.Lucarelli M, Narzi L, Pierandrei S, Bruno SM, Stamato A, d’Avanzo M, et al. A new complex allele of the CFTR gene partially explains the variable phenotype of the L997F mutation. Genet Med. 2010;12:548–55. doi: 10.1097/GIM.0b013e3181ead634. [DOI] [PubMed] [Google Scholar]
- 10.Romey MC, Guittard C, Chazalette JP, Frossard P, Dawson KP, Patton MA, et al. Complex allele [-102T>A+S549R(T>G)] is associated with milder forms of cystic fibrosis than allele S549R(T>G) alone. Hum Genet. 1999;105:145–50. doi: 10.1007/s004399900066. [DOI] [PubMed] [Google Scholar]
- 11.Salles MV, Motta FL, Dias da Silva E, Varela P, Costa KA, Filippelli-Silva R, et al. Novel Complex ABCA4 Alleles in Brazilian Patients With Stargardt Disease: Genotype-Phenotype Correlation. Invest Ophthalmol Vis Sci. 2017;58:5723–30. doi: 10.1167/iovs.17-22398. [DOI] [PubMed] [Google Scholar]
- 12.Shroyer NF, Lewis RA, Lupski JR. Complex inheritance of ABCR mutations in Stargardt disease: linkage disequilibrium, complex alleles, and pseudodominance. Hum Genet. 2000;106:244–8. doi: 10.1007/s004399900222. [DOI] [PubMed] [Google Scholar]
- 13.Zaum AK, Stuve B, Gehrig A, Kolbel H, Schara U, Kress W, et al. Deep intronic variants introduce DMD pseudoexon in patient with muscular dystrophy. Neuromuscul Disord. 2017;27:631–4. doi: 10.1016/j.nmd.2017.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.