Abstract
Background:
Alcohol use disorder (AUD) and schizophrenia (SCZ) frequently co-occur, and large-scale genome-wide association studies (GWAS) have identified significant genetic correlations between them.
Methods:
We used the largest published GWAS for AUD (total cases = 77,822) and SCZ (total cases = 46,827) to identify genetic variants that influence both disorders (with either the same or opposite direction of effect) and those that are disorder specific.
Results:
We identified 55 independent genome-wide significant SNPs with the same direction of effect on AUD and SCZ, 8 with robust effects in opposite directions, and 98 with disorder-specific effects. We also found evidence for 12 genes whose pleiotropic associations with AUD and SCZ are consistent with mediation via gene expression in the prefrontal cortex. The genetic covariance between AUD and SCZ was concentrated in genomic regions functional in brain tissues (p = 0.001).
Conclusions:
Our findings provide further evidence that SCZ shares meaningful genetic overlap with AUD.
Introduction
Schizophrenia (SCZ) and alcohol dependence or abuse (hereafter referred to as alcohol use disorder, AUD) are serious psychiatric disorders(Whiteford et al., 2013). AUD is more common in individuals with SCZ (prevalence of 20–30%(Castillo-Carniglia, Keyes, Hasin, & Cerdá, 2019; Hunt, Large, Cleary, Lai, & Saunders, 2018), compared to ~6% in the general population(SAMHSA, 2019)) and a diagnosis of both is associated with greater psychiatric comorbidity(Brady, Killeen, & Jarrell, 1993), more clinical complications(Duke, Pantelis, & Barnes, 1994), and a lower likelihood of sustained medication adherence(Drake & Wallach, 1989) than either disorder alone. Both SCZ and AUD are moderately to highly heritable (twin-h2 for SCZ = 81%(PF, KS, & MC, 2003), AUD = 49%(Verhulst, Neale, & Kendler, 2015)), and genome-wide association studies (GWAS) have consistently found positive genetic correlations of AUD with SCZ (e.g., rg = 0.34, p = 3.7e-21)(Kranzler et al., 2019), suggesting that common genetic influences may potentially contribute to their co-occurrence (although environmental factors undoubtedly play a large role). Further, polygenic risk scores (PRS) for AUD are significantly associated with SCZ risk(Kranzler et al., 2019) and vice versa(Reginsson et al., 2017). In contrast, the genetic correlation between SCZ and measures of typical alcohol consumption is weak (e.g., drinks/week: rg = 0.01, p = 0.67)(Liu et al., 2019), suggesting that SCZ might share substantial genetic liability only with the psychopathological aspects of disordered drinking(Sanchez-Roige et al., 2018) and not alcohol consumption per se.
Despite the substantial genetic correlation between AUD and SCZ, there is little known regarding the underlying pleiotropic mechanisms in terms of the specific risk alleles, genes, and molecular pathways involved. Although recent studies have begun to elucidate the contributions of pleiotropic loci to shared genetic variance amongst disorders and complex traits(Lam et al., 2019; Lee et al., 2019), these efforts have not included AUD, one of the most common psychiatric disorders; until recently, GWAS of AUD were not well-powered. In addition to loci with a similar direction of effect on both disorders, modern cross-disorder GWAS methods can also identify divergent variants, i.e., those that are pleiotropic for two disorders but whose effect alleles operate in opposite directions, conferring risk for one disorder and protective effects for the other (e.g., potassium ion response genes that distinguish SCZ from bipolar disorder(Ruderfer et al., 2018)). The identification of such variants is fundamental to identifying the pathways that contribute to diagnostic boundaries.
The current study outlines the nature of the shared genetic underpinnings of AUD and SCZ by conducting cross-disorder analyses of large genome-wide datasets of both European- and African-ancestry individuals (see Figure 1 for overview). We conducted ancestry-specific cross-disorder meta-analyses to systematically identify pleiotropic loci with significant convergent and divergent effects on both SCZ and AUD, and loci specific to each disorder. We also linked pleiotropic variants to gene expression data from the frontal cortex in an effort to prioritize the genes that are more likely to be causal. Because the extent to which the correlation between AUD and SCZ is attributable to salient functional categories remains unknown, we partitioned the genetic covariance between AUD and SCZ into relevant annotations.
Figure 1.
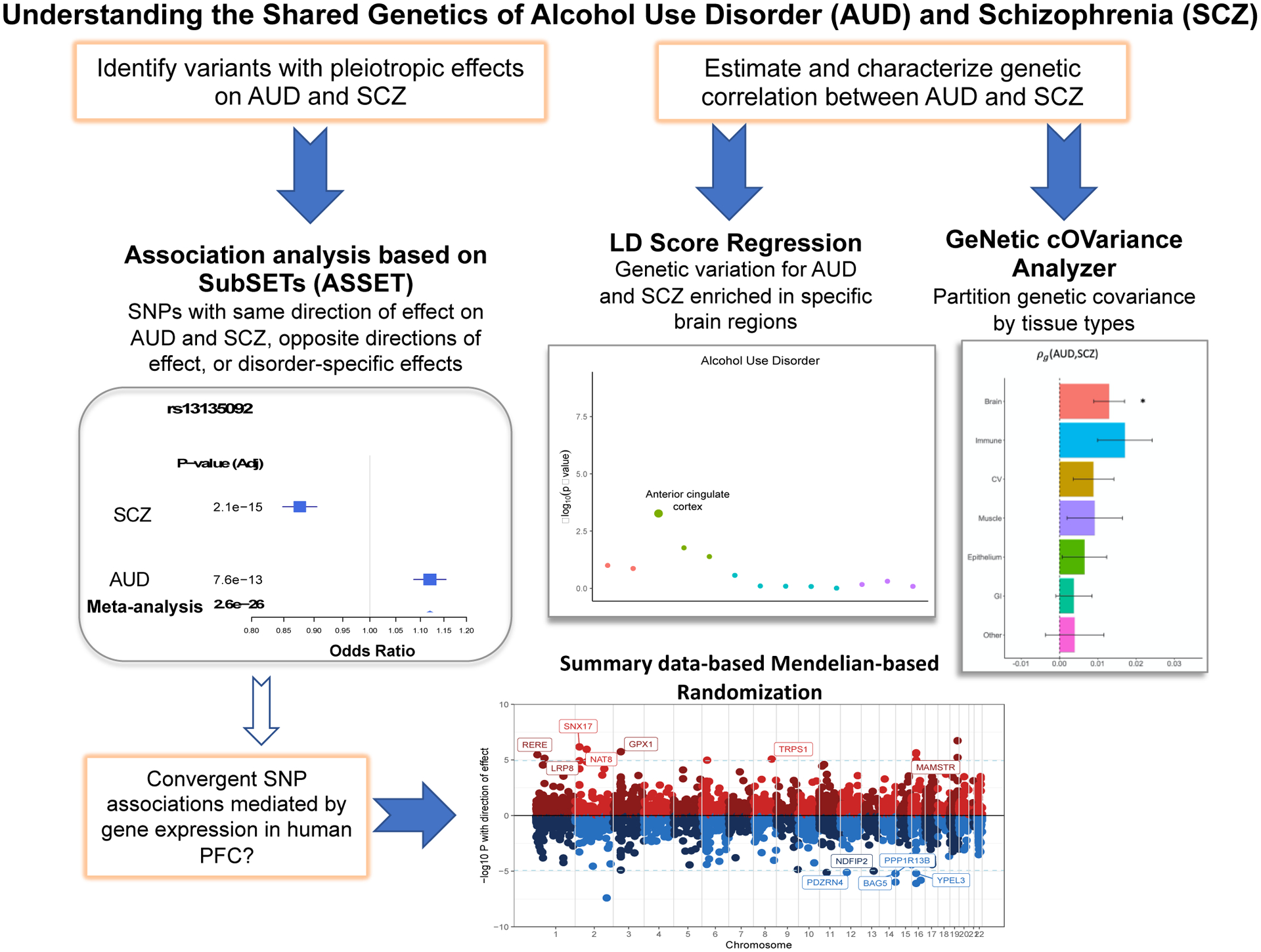
Conceptual overview of cross-disorder analysis of alcohol use disorder and schizophrenia.
Methods and Materials
Samples
Alcohol Use Disorder
For the European-ancestry subset, we meta-analyzed a subset of the GWAS data from the largest available AUD meta-analysis(Zhou, Sealock, et al., 2020) (N = 313,959; Ncases = 57,564): we meta-analyzed two GWAS of alcohol dependence and abuse (MVP + PGC), and did not include the GWAS of the problem subscale of the Alcohol Use Disorders Identification Test in the UK Biobank. In the Million Veteran Program (MVP), case status was derived from International Classification of Diseases (ICD) codes of alcohol dependence and abuse in electronic health records (EHR). In the Psychiatric Genomics Consortium (PGC) alcohol dependence GWAS(Walters et al., 2018), cases were defined by DSM-IV diagnoses.
We also meta-analyzed two published GWAS to create the African-ancestry subset: the MVP Phase 1 GWAS of AUD(Kranzler et al., 2019) (N = 56,648; Ncases = 17,267), and the PGC GWAS of alcohol dependence(Walters et al., 2018) (N = 5,799; Ncases = 2,991). We used METAL(Willer, Li, & Abecasis, 2010) (which combines genome-wide summary statistics across multiple samples) to generate the African-ancestry summary statistics by meta-analyzing the GWAS data from the MVP Phase 1 AUD and PGC alcohol dependence GWAS using an inverse variance-weighted fixed-effects model, excluding SNPs with INFO score < 0.8 and/or minor allele frequency < 0.01 within each sample.
Schizophrenia
For the European-ancestry sample, we used the PGC Phase 2 + CLOZUK (a sample of individuals with schizophrenia who were treated with clozapine) SCZ GWAS meta-analysis(Pardiñas et al., 2018) (total N = 105,318; Ncases = 40,675). We used the summary statistics from the Genomic Psychiatry Cohort (GPC) SCZ GWAS(Bigdeli et al., 2019) (N = 10,070; Ncases = 6,152) for the African-ancestry cross-disorder analysis.
Analysis
Cross-disorder association analysis
We used “Association analysis based on SubSETs” (ASSET)(Bhattacharjee et al., 2012) to combine the genome-wide association data for AUD and SCZ (separately by ancestry), using the two-tailed meta-analysis approach to obtain a single cross-disorder association statistic, correcting for sample overlap. Unlike traditional meta-analysis approaches, ASSET takes into account SNPs with significant effects on multiple disorders even if the effects on the traits are in opposite directions. Default parameters were applied using the “h.traits” function, and we used LD Score Regression(B. Bulik-Sullivan et al., 2015; B. K. Bulik-Sullivan et al., 2015) (LDSC) to obtain a rough estimate of sample overlap, which we accounted for in the additional covariance term. We then separated the ASSET results into four subsets: a “convergent” subset (effect allele with the same direction of effect for both disorders), a “divergent” subset (effect allele with opposite directions of effect on the disorders), a subset of SNPs with AUD-specific effects, and a subset of SNPs with effects only on SCZ (Supplemental Figure 1).
In the European-ancestry sample, we then uploaded the subset results (i.e., convergent, divergent, AUD-only, and SCZ-only SNPs from ASSET output) to FUMA v1.3.6a(Watanabe, Taskesen, van Bochoven, & Posthuma, 2017) for annotation and identification of genome-wide significant risk loci and independent lead SNPs. We further subset these convergent and divergent loci to exclude top lead SNPs with p > 0.05 in either individual disorder GWAS, to create a more conservative set of cross-disorder variants with at least nominal significance in both disorders (although the full subsets were used for gene-set and pathway analyses).
In the African-ancestry samples, which were smaller and accordingly lacked power for more extensive analyses, we focused on the overall set of pleiotropic cross-disorder variants, rather than parsing the pleiotropic variants into subsets with convergent and divergent effects.
To examine specificity of genetic overlap, we also examined the effect sizes and p-values of the top pleiotropic loci identified for AUD and SCZ in GWAS of attention deficit hyperactivity disorder (ADHD)(Demontis et al., 2019), bipolar disorder(Stahl et al., 2019), depression(Howard et al., 2019), cigarettes per day(Liu et al., 2019), cannabis use disorder(Johnson et al., 2020), and opioid use disorder(Zhou, Rentsch, et al., 2020).
Gene, gene-set, and pathway analyses
In both the European-ancestry and African-ancestry samples, gene-based analyses in MAGMA (v1.08)(de Leeuw, Mooij, Heskes, & Posthuma, 2015) were conducted on the subset results (i.e., convergent and divergent SNPs from ASSET output) via the FUMA(Watanabe et al., 2017) platform. These analyses included gene-set analyses using curated gene sets and GO terms from MsigDB(Liberzon et al., 2011) (an online collection of annotated gene-sets) and gene-property analyses based on tissue expression data from GTEx(Aguet et al., 2017) v8 (a repository of expression data by brain region from autopsies of 960 donors) and brain samples at different developmental stages and specific ages from BrainSpan(Miller et al., 2014) (more details in Supplemental Materials).
Summary data-based eQTL analyses
To examine whether the effects of pleiotropic variants with convergent effects on AUD and SCZ may be mediated by gene expression patterns, we conducted a summary data-based Mendelian randomization(Zhu et al., 2016) (SMR) analysis on a set of expression quantitative trait loci (eQTL) data in the prefrontal cortex (meta-analyzed to combine data from ROSMAP(De Jager et al., 2018), PsychENCODE(Akbarian et al., 2015), and COGA-INIA datasets(Kapoor et al., 2019); total N = 1,986). SMR is a Mendelian randomization-based analysis that integrates GWAS summary statistics with eQTL data to test whether the effect size of a SNP on the phenotype of interest is mediated by gene expression. SMR does not require raw eQTL data to build the weights. We excluded variants with pleiotropic effects significantly different from what would be expected under a causal model using the HEIDI-outlier method(Zhu et al., 2018) (excluding SNPs with HEIDI-outlier p < 0.05).
Differential gene expression analyses using AUD and SCZ post-mortem samples
We also examined whether genes mapped to pleiotropic loci by MAGMA (with p < 0.05) were significantly enriched for genes showing differential expression (p < 0.05) in the prefrontal cortex using two comparisons in independent samples: we compared gene expression in 65 individuals with alcohol dependence and 73 healthy controls(Kapoor et al., 2019), and 258 individuals with SCZ and 279 controls(Fromer et al., 2016) (details in Supplemental Note) using Fisher’s exact test(Fisher, 1934).
Genetic correlations and partitioned covariance
We used LDSC(B. Bulik-Sullivan et al., 2015; B. K. Bulik-Sullivan et al., 2015) to estimate the genetic correlations (rg) between AUD, SCZ, and two negative control traits (height and chronic ischemic heart disease). We compared the genetic correlations between AUD and SCZ, and DPW and SCZ using a block jackknife method implemented through LDSC to test the null hypothesis that rg(AUD, SCZ) minus rg(DPW, SCZ) = 0.
We also used LDSC applied to Specifically Expressed Genes (LDSC-SEG(Finucane et al., 2018)) to estimate the enrichment of AUD and SCZ across 13 specific brain regions (annotations defined using GTEx(Lonsdale et al., 2013) gene expression data).
We used GeNetic cOVariance Analyzer (GNOVA)(Lu, Li, et al., 2017) to partition the genetic covariance (ρg) between AUD and SCZ into salient annotation categories. These included 1) functional vs. non-functional areas of the genome (GenoCanyon(Lu et al., 2015) annotations, defined by integrating genomic conservation measures and biochemical annotation data to generate a functional potential score for each genetic variant), 2) tissue- and regional-specific functionality (GenoSkyline(Lu, Powles, et al., 2017; Lu, Powles, Wang, He, & Zhao, 2016) annotations, which are tissue-specific functional regions defined by integrating high-throughput epigenetic annotations, 3) GTEx v6(Lonsdale et al., 2013) brain region annotations (adapted from LDSC cell-type specific analyses), and 4) minor allele frequency quartiles. We excluded the MHC region (chr6:26000885 - chr6:33999991) from both the LDSC-SEG and GNOVA analyses due to the long-range and complex LD in this region.
In sensitivity analyses conducted on individuals of European ancestry in the UK Biobank (from the Neale lab GWAS: https://www.nealelab.is/uk-biobank), we calculated the genetic correlation and partitioned genetic covariance between AUD, SCZ, and two negative control traits: height (N = 360,388) and chronic ischemic heart disease (CHD; N = 361,194; Ncases = 12,769).
Results
Identifying pleiotropic variants, genes, and pathways in individuals of European ancestry
The cross-disorder analysis of AUD and SCZ in European-ancestry individuals identified numerous pleiotropic loci: after genome-wide clumping via FUMA(Watanabe et al., 2017), there were 55 independent risk loci (with 60 lead SNPs) with convergent effects and 44 risk loci (56 lead SNPs) with divergent effects (i.e., effect allele with opposite effects on AUD and SCZ; Supplemental Figure 1, Supplemental Tables 1–3). We also identified disorder-specific loci through ASSET: 90 with SCZ-only effects, and 8 with AUD-only effects (Supplemental Tables 4–5). MAGMA gene-based analyses identified 119 significant genes from the convergent subset and 105 genes from the divergent subset (Supplemental Figure 2, Supplemental Tables 6–7).
As ASSET(Bhattacharjee et al., 2012) searches for and determines the most likely subset for each SNP (i.e., classifying SNPs as having an effect only on AUD, only on SCZ, or on both disorders), some of the pleiotropic SNPs identified by ASSET were only significant in one of the single-disorder GWAS. For a more conservative description of pleiotropic loci of divergent effect, we further considered only the 8 loci where the top lead SNPs had opposite effects but p < 0.05 for both AUD and SCZ (Supplemental Table 3; the convergent lead SNPs already had p < 0.05 for both disorders). In the convergent subset, the strongest association was on chromosome 11 (lead SNP rs6589386, cross-disorder p = 5.7e-18; AUD p = 7.1e-12, SCZ p = 1.6e-8). The strongest divergent signal was on chromosome 4 (lead SNP rs13135092, cross-disorder p = 2.9e-31; AUD p = 4.9e-18, SCZ p = 7.9e-16), located in an intron of SLC39A8; the effect allele (A) increases risk for AUD and decreases risk (i.e., is protective) for SCZ.
MAGMA competitive gene-set analyses identified 2 significant GO terms in the convergent subset of variants, one related to DNA binding (GO: 0043565, p = 1.3e-6) and one related to neuronal differentiation (GO: 0045664, p = 1.9e-6; Supplemental Table 8). There were no significant gene-sets identified for the divergent subset (Supplemental Table 9).
Both the convergent and divergent subsets of variants showed enrichment in all 13 brain tissues in MAGMA gene-property analyses, and the divergent subset also showed enrichment in pituitary tissues. (Supplemental Figure 3). The convergent subset of variants showed enrichment for the early-mid prenatal general developmental stage in the BrainSpan data, but did not show any significant enrichments in the 29 more specific age samples (Supplemental Figure 3). The divergent subset did not show enrichment in any of the specific ages or developmental stages.
Cross-disorder variants, genes, and pathways in African ancestry individuals
In the African-ancestry cross-disorder analysis, there was limited power to identify pleiotropic loci, with results appearing to be primarily driven by the larger AUD GWAS (e.g., the one genome-wide significant SNP (rs150627184) that ASSET identified as being pleiotropic had p > 0.05 for SCZ; Supplemental Table 10). There were three significant genes (ADH4, ADH1B, and EIF4E) identified in the MAGMA gene-based analysis (Supplemental Figure 4 and Supplemental Table 11). No gene-sets passed Bonferroni correction in the competitive gene-set analysis (Supplemental Table 12), and gene-property analyses did not reveal significant enrichment for any tissues or developmental stages (Supplemental Figure 5). While neither of the two specific gene-sets identified in the European-ancestry cross-disorder analysis (related to DNA binding and neuron differentiation) were significant in the African ancestry analysis after multiple testing corrections, a gene-set related to the regulation of dopaminergic neuron differentiation had p = 0.003.
Overlap of top loci with other psychiatric and substance use GWAS
To assess the specificity of the identified divergent and convergent loci, we examined the effect sizes of these loci in several European-ancestry psychiatric and substance use GWAS (Supplemental Tables 13 & 14): ADHD, bipolar disorder, depression, cigarettes per day, cannabis use disorder, and opioid use disorder. Of the top lead SNPs at the 55 convergent loci, 22 showed statistically significant effects (Bonferroni corrected for the 55 SNPs tested: α = 0.0009) across one or more of the six phenotypes tested. Depression shared the most convergent SNPs with AUD and SCZ, with 10 of the 55 lead SNPs having p < 0.0009 in the depression GWAS. In the subset of 8 divergent SNPs with p < 0.05 in both AUD and SCZ, 3 were significantly associated (α = 0.00625) in one or more of the other GWAS. In particular, cigarettes per day showed a strong association (p = 1.2e-123) with rs8042374, an intronic variant in the CHRNA3 gene which exerts an effect in the same direction for both the AUD and cigarettes per day GWAS and in the opposite direction of effect for the SCZ GWAS.
Genetically regulated gene expression analyses
Of the 22 genes that survived Bonferroni correction (for the number of genes tested; pSMR < 1.16e-5), SMR analyses in the European ancestry sample identified 12 convergent genes whose cross-disorder associations with AUD and SCZ were consistent with mediation via gene expression in the prefrontal cortex (PFC; the remaining 10 genes with pSMR < 1.16e-5 had pHEIDI < 0.05, indicating pleiotropy outlier status; Figure 2; significant results in Supplemental Table 15; full results in Supplemental Table 16).
Figure 2. Genes in the convergent subset whose association with AUD and SCZ may be mediated by gene expression in the prefrontal cortex, analyzed using Summary data-based Mendelian Randomization (SMR).
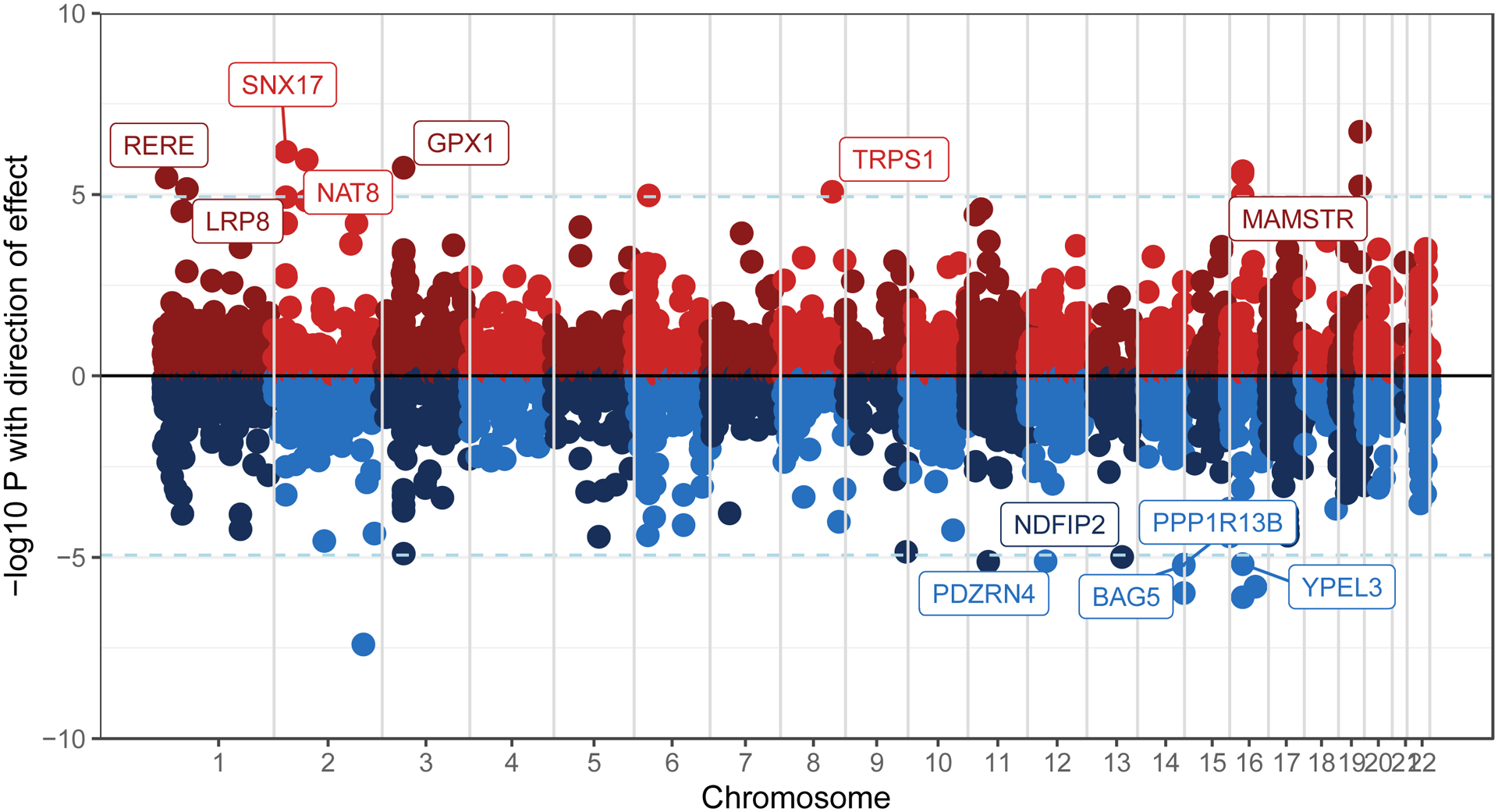
Genes in red show up-regulated gene expression, and genes in blue show down-regulated gene expression. The 12 labeled genes are significant after Bonferroni corrections and were not excluded as pleiotropic outliers (HEIDI-outlier method p > 0.05)
Differential gene expression enrichment analyses
Neither the convergent nor divergent genes identified in the European ancestry sample were enriched in differential gene expression analyses of postmortem PFC tissue of individuals with SCZ (Ncases = 258) vs. controls or of AUD (Ncases = 65) vs. controls.
Genetic covariance and correlation
There were significant positive genetic correlations (rg) between AUD and SCZ (rg = 0.392, p = 1.2e-42). In sensitivity tests, the negative control measure of chronic ischemic heart disease was not significantly correlated with AUD but height showed a small negative correlation (rg = −0.093, SE = 0.021, p = 7.54e-6). SCZ showed a nominal genetic correlation with heart disease (rg = −0.063, SE = 0.029, p = 0.032), but none with height.
LDSC-SEG(Finucane et al., 2018) analyses that partitioned heritability enrichment by tissue-specific gene expression revealed significant enrichment for SCZ in three of 13 brain regions: the cortex, frontal cortex, and anterior cingulate cortex (FDR q-values < 2e-5), and for AUD in the anterior cingulate cortex (FDR q-value = 0.007; Supplemental Figure 6).
The genetic covariance (ρg) of AUD and SCZ was significantly attributable both to functional regions of the genome (p = 1.0e-8), including those specifically functional in brain tissues (p = 0.001; see Figure 3a, Supplemental Table 17) and non-functional regions (p = 1.5e-19; Supplemental Table 18), as well as all minor allele frequency quartiles except the lowest frequency quartile (p = 2.9e-5 – 3.9e-9; Supplemental Table 19). While the point estimate of genetic covariance was highest in genes functional in immune tissues (ρg = 0.017), this estimate had a relatively large standard error (0.007) and did not reach significance after multiple testing corrections. There were no significant findings when partitioning the genetic covariance between AUD and height or SCZ and heart disease into tissue-specific categories with GNOVA.
Figure 3. Stratified genetic covariance between AUD and SCZ.
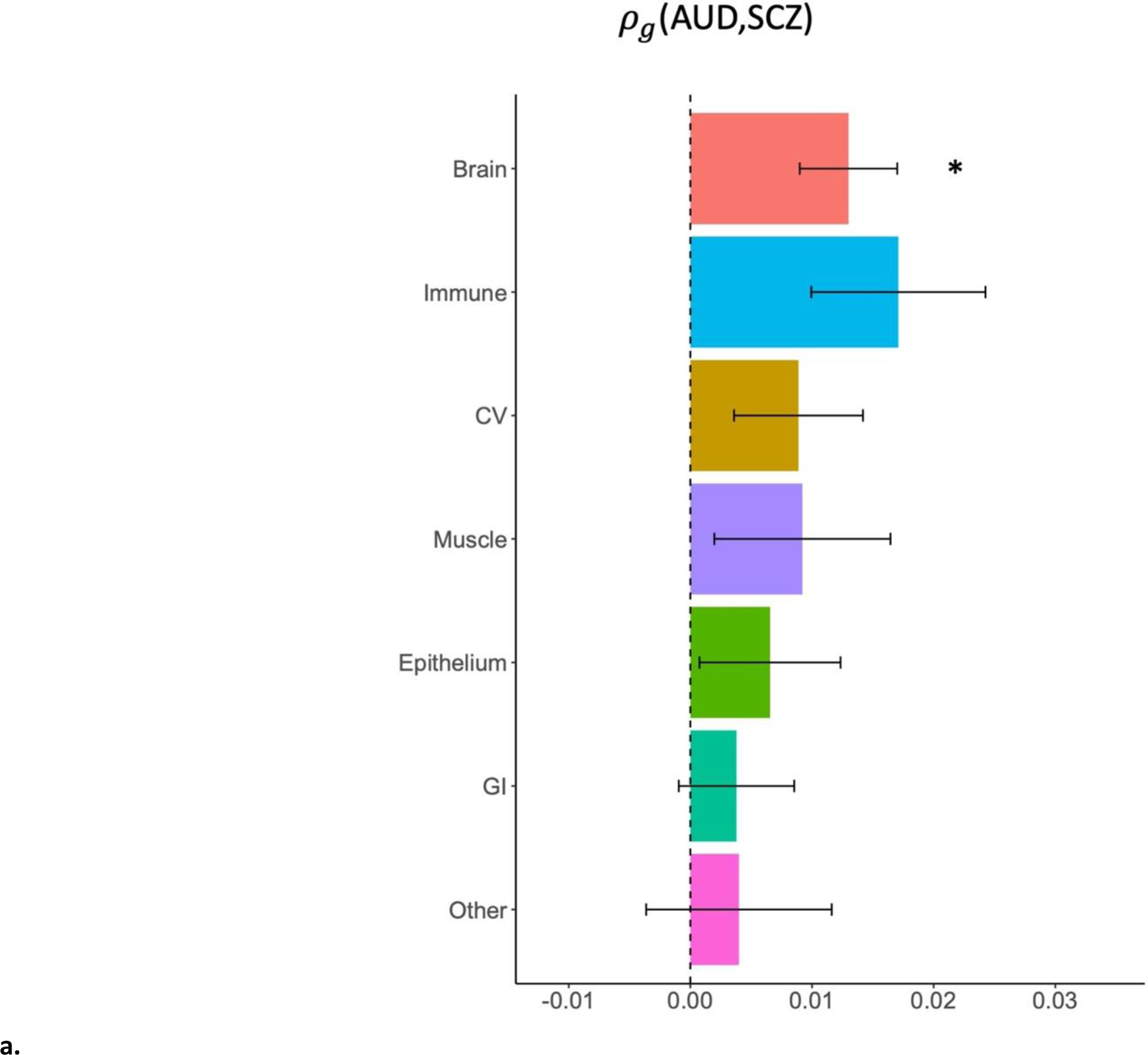
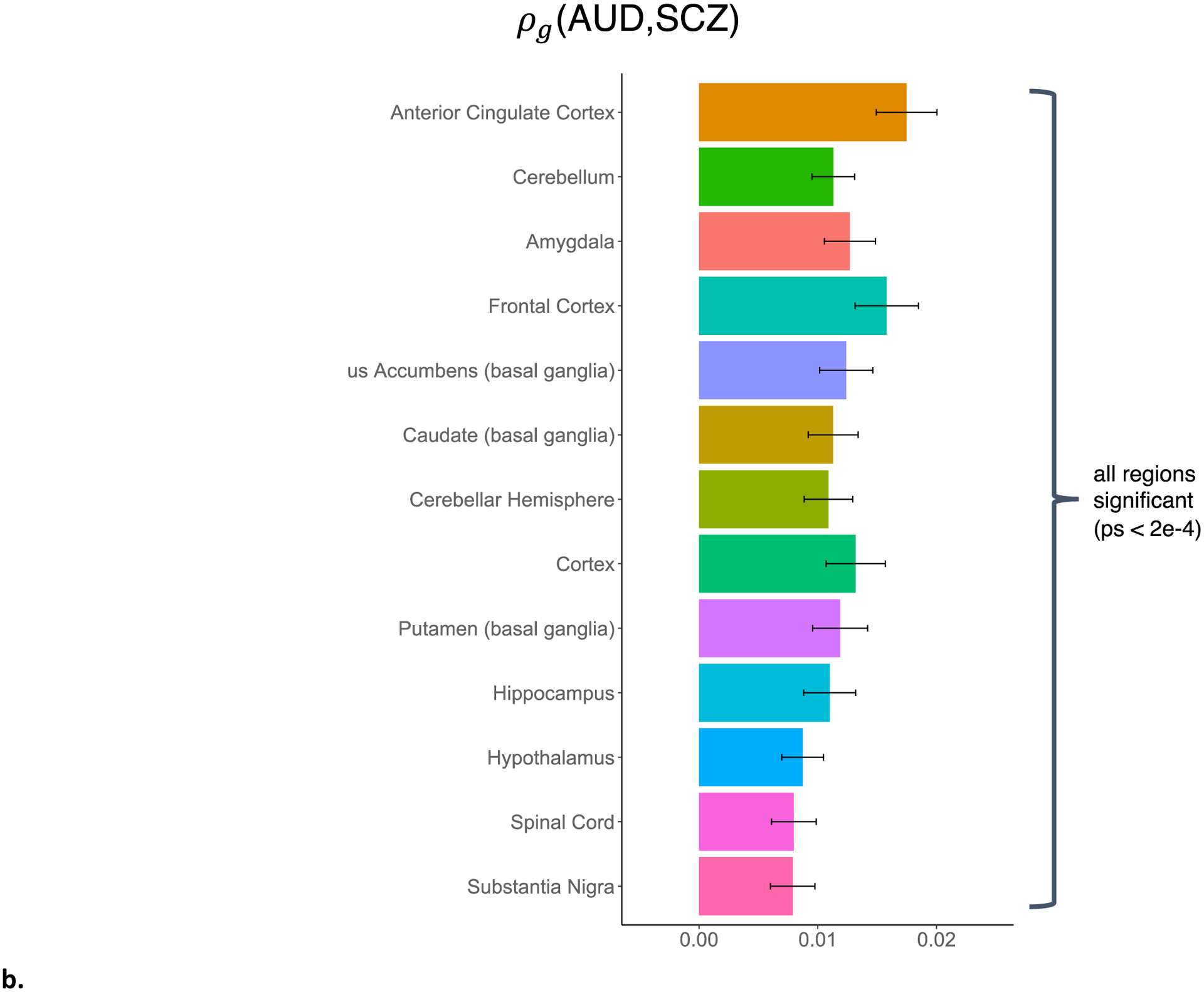
a: stratified by broad tissue type. Tissue annotations were defined using the GenoSkyline-Plus annotations. Significant tissues are starred. CV = cardiovascular; GI = gastrointestinal. b: stratified by 13 brain regions, defined using GTEx v6 gene expression data.
Because the genetic covariance of AUD and SCZ was significantly concentrated in brain tissues, we partitioned it further into 13 specific brain regions. We found significant concentrations of genetic covariance in all 13 brain regions tested, with the greatest concentrations in the anterior cingulate cortex (p = 7.6e-12), frontal cortex (p = 3.3e-9), cortex (p = 1.2e-7), and amygdala (p = 3.1e-9; Supplemental Table 20, Figure 3b).
Discussion
The prevalence of AUD is elevated in those with SCZ, relative to the general population. Although environmental factors likely play a large role in this comorbidity, there is some evidence that shared genetic influences may also contribute, with recent large GWAS of AUD documenting robust genetic correlations between AUD and SCZ(Zhou, Sealock, et al., 2020). Utilizing a subset-based meta-analytic approach, our cross-disorder analysis of AUD and SCZ identified 55 convergent loci with the same direction of effect on AUD and SCZ and 8 divergent loci with opposite directions of effect that had lead SNPs with p < 0.05 in both individual disorders. The genetic covariance between AUD and SCZ was concentrated in genes functional in the brain, and eQTL analyses of the convergent subset of variants identified 12 genes whose association with AUD and SCZ may be mediated via gene expression in brain tissues.
In contrast to the robust genetic correlation between AUD and SCZ, prior studies have found that SCZ is uncorrelated with measures of typical alcohol consumption (e.g., with drinks/week, rg = 0.01, p = 0.67(Liu et al., 2019)) or frequency of use(Marees et al., 2019). Genetic correlations with indices that encompass heavy episodic drinking, such as the consumption sub-scale of the Alcohol Use Disorders Identification Test (AUDIT-C), are also lower (rg = −0.0003 −0.04)(Kranzler et al., 2019; Sanchez-Roige et al., 2018) than those noted for AUD. A recent study suggests that indices of socioeconomic status may influence genetic correlations between measures of substance use and psychopathology(Marees et al., 2020); however, even after co-varying for SES, the genetic correlation between alcohol frequency and SCZ remained non-significant. The genetic correlation between drinks per week and SCZ remained significant and relatively unchanged (rg = 0.14–0.16), but markedly lower than the AUD-SCZ genetic correlation. This suggests that SCZ shares genetic variation primarily with the psychopathological aspects of problem drinking and AUD. Future studies that employ symptom-level GWAS, or GWAS of clinically relevant phenotypes (e.g., negative symptoms of SCZ), may yield further insights into whether this genetic overlap is specific to certain symptoms or aspects of these psychiatric disorders.
The top divergent SNP, rs13135092, in an intron of SLC39A8, was strongly associated with both AUD (p = 4.9e-18) and SCZ (p = 7.9e-16), with the A allele exerting a risk-increasing effect on AUD and a protective effect on SCZ. Comparison of summary statistics for over 4,756 traits(Watanabe et al., 2019) indicated that the A allele was also associated with greater alcohol consumption, greater risk-taking, higher waist-hip ratio, lower bioelectrical impedance (i.e., greater adiposity) and higher systolic blood pressure, consistent with the direction of genetic association between these measures and AUD. However, the A allele was also associated with increasing cognitive performance, higher intelligence and with higher educational attainment – while the direction of these effects is consistent with the “protective” effect of the A allele on risk for SCZ, it contradicts prior research showing an inverse genetic correlation between educational attainment and AUD. Given the convergent direction of associations with AUD and alcohol consumption, risk-taking, and cardio-metabolic traits, but divergent direction of associations with SCZ and cognition, we speculate that the A allele of rs13135092 may be related to milder AUD, typified by positive reward-related drinking and impulsivity that is effectively regulated by enhanced cognitive functioning.
Our top convergent SNP, rs6589386, is intergenic and is an eQTL for DRD2 in cerebellar hemisphere tissue; DRD2 has been implicated in both AUD(Kranzler et al., 2019) and SCZ(Consortium, 2014) GWAS. In addition to AUD and SCZ, this variant has been implicated in GWAS of neuroticism(Baselmans et al., 2019), subjective well-being(Baselmans et al., 2019), alcohol consumption(Liu et al., 2019), and cigarette smoking(Liu et al., 2019).
Consistent with recent reports demonstrating broad levels of pleiotropy amongst psychiatric disorders(Lee et al., 2019), including substance use disorders(Hatoum et al., 2021), we found that 22 of the 55 top convergent SNPs and 3 of the 8 divergent SNPs showed at least nominal significance across one or more of the six psychiatric disorders and substance use traits that we assessed. Depression showed the greatest extent of overlap with the convergent SNPs, with 10 of the 55 convergent SNPs having p < 0.0009 in the depression GWAS and only one of those exerting a different direction of effect relative to AUD and SCZ. Of the 8 top SNPs in the divergent subset, both cigarettes per day and opioid use disorder were significantly (p < 0.00625) associated with two of those SNPs, with both variants showing the same direction of effect as for AUD (and opposite direction of effect from SCZ), again consistent with the pleiotropy observed among different substance use traits. While there may be individual loci and pathways that contribute uniquely to AUD and SCZ, it is evident that much of this genetic overlap may also be shared with other psychiatric disorders and substance use traits.
SMR eQTL analyses suggest that the effects of pleiotropic variants on AUD and SCZ may be mediated by expression levels of several genes in the prefrontal cortex (PFC; Figure 2). Several of the identified genes were previously implicated in GWAS of metabolic traits (including NAT8, TRPS1 (up-regulated), BAG5, PPP1R13B (down-regulated)), immunological traits (e.g., RERE, TRPS1, both up-regulated) and psychiatric phenotypes (e.g., up-regulated: LRP8, down-regulated: PPP1R13B). The data currently available do not permit examination of whether these findings extend to other brain regions.
We found no evidence for enrichment of genes pleiotropically associated with both AUD and SCZ when considering genes differentially expressed in the brains of individuals with AUD or SCZ (compared to respective controls). Unlike the SMR eQTL analysis described above, which reflects the genetically regulated portion of gene expression, the differential gene expression measured in autopsy samples likely reflects pathological consequences of the disorders (AUD and SCZ) and their treatments. However, our lack of findings could be due to limited statistical power in the RNAseq whole-genome transcriptome datasets, especially the post-mortem sample of individuals with AUD (Ncases = 65).
There are several limitations of the current study. First, nearly all GWAS, gene expression reference datasets, and other bioinformatic resources are based on European-ancestry samples. Our African-ancestry GWAS samples were under-powered relative to the European-ancestry samples, and the SCZ sample more so than the AUD sample. Likely due to the statistical power differential, the African-ancestry results appeared to be driven almost entirely by the larger AUD GWAS. One of our motivations for conducting this cross-disorder analysis is to reduce inequity in downstream analyses of non-European populations; the genome-wide summary statistics from our cross-disorder GWAS could be used to generate polygenic risk scores in independent cohorts of African-ancestry participants. Our findings underscore the need for vastly larger African-ancestry samples for studies of severe psychiatric conditions. Second, given the available data, we were limited to studying common genetic variation (minor allele frequencies > 1%); thus, there may be rare variants of importance underlying the comorbidity between AUD and SCZ that are outside the scope of the current study. Third, in our sensitivity analysis, we found a significant negative genetic correlation between AUD and height (rg = −0.093, SE = 0.021, p = 7.54e-6), which is likely driven in part by the large sample sizes and polygenic nature of both traits, though there is some evidence for this relationship in the literature. One previous study found a negative genetic correlation between alcohol consumption and childhood height in males(Clarke et al., 2017) (rg = −0.23, p = 0.002) and another study found a negative correlation between problematic alcohol use and comparative height at age 10 (rg = −0.09, p = 7.3e-5), though this was not significant when a Bonferroni correction was applied for the 715 traits tested(Zhou, Sealock, et al., 2020). Another limitation of our cross-sectional design is that we are unable to establish whether shared genetic influences, as identified in this and previous studies, are causal factors for the comorbidity of AUD and SCZ. While we focused exclusively on genetic factors that influence the risk of AUD and SCZ, social and environmental factors are likely to play at least as large a role as genetic ones in the development and co-occurrence of these disorders, and our findings should be interpreted in light of this. Finally, while most of the AUD samples were screened for SCZ, some individuals among the SCZ samples very likely also had AUD. These potential cases of unknown comorbidity could have biased our estimates of genetic correlation and pleiotropic loci. Unfortunately, there are no data currently available on which SCZ samples were screened for substance use disorders to permit an estimation of such a bias.
While prior cross-disorder studies have provided foundational results for common psychiatric disorders, ours is amongst the first studies to focus on identifying pleiotropic loci for a substance use disorder, AUD, and SCZ. To understand the shared biology between substance use disorders and other psychiatric disorders, future efforts must include large numbers of individuals with AUD or problematic alcohol use rather than simple measures of consumption.
Supplementary Material
ACKNOWLEDGEMENTS
The Psychiatric Genomics Consortium’s Substance Use Disorders (PGC-SUD) working group gratefully acknowledges prior support from NIAAA and thanks all our contributing investigators and study participants who make this research possible. We acknowledge collaborations with the Million Veteran Program, Psychiatric Genomics Consortium Substance Use Disorders Working Group, and Genomic Psychiatry Cohort Investigators. A list of Genomic Psychiatry Cohort investigators and the MVP Core Acknowledgement are provided in the Supplemental Materials. We are grateful to Luke Evans for helpful discussion of the manuscript.
FUNDING
ECJ was supported by F32AA027435. RP was supported by R21DA047527 and R21DC018098. FRW was supported by F32MH122058. AA is supported by K02DA032573. REP is supported by NIMH K01MH113848 and The Brain & Behavior Research Foundation NARSAD grant 28632 P&S Fund.
This work was conducted using the summary statistics from the Psychiatric Genomics Consortium’s Substance Use Disorders Working group. The Psychiatric Genomics Consortium’s Substance Use Disorders (PGC-SUD) working group is supported by MH109532 with funding from NIMH and NIDA.
The GPC was supported by grants R01 MH085548 and R01 MH104964 from the National Institute of Mental Health (NIMH), and genotyping of samples was provided by the Stanley Center for Psychiatric Research at Broad Institute.
COGS was supported by grants R01 MH065571, R01 MH065588, R01 MH065562, R01 MH065707, R01 MH065554, R01 MH065578, R01 MH065558, R01 MH86135, and R01 MH094320 from the National Institute of Mental Health.
Funding support for the Whole Genome Association Study of Bipolar Disorder and the Genome-Wide Association of Schizophrenia Study was provided by the NIMH (R01 MH67257, R01 MH59588, R01 MH59571, R01 MH59565, R01 MH59587, R01 MH60870, R01 MH59566, R01 MH59586, R01 MH61675, R01 MH60879, R01 MH81800, U01 MH46276, U01 MH46289, U01 MH46318, U01 MH79469, and U01 MH79470) and the genotyping of samples was provided through the Genetic Association Information Network (GAIN).
This research is based, in part, on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, with support from award #I01 BX003341 and the VISN 4 Mental Illness Research, Education and Clinical Center. This publication does not represent the views of the Department of Veterans Affairs or the United States Government.
FINANCIAL DISCLOSURES
Dr. Kranzler is a member of an advisory board for Dicerna Pharmaceuticals, a consultant for Sophrosyne Pharmaceuticals, and a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last three years by AbbVie, Alkermes, Dicerna, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor, and Amygdala Neurosciences. Dr. Kranzler and Dr. Gelernter are named as inventors on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018. Drs. Gelernter and Polimanti are paid for their editorial work on the journal Complex Psychiatry. Drs. Johnson, Kapoor, Hatoum, Zhou, Wendt, R.K. Walters, Lai, Kember, Hartz, Meyers, Peterson, Ripke, Bigdeli, Fanous, C.N. Pato, M.T. Pato, Goate, O’Donovan, J.T.R. Walters, Edenberg, and Agrawal have no disclosures or conflicts of interest to report.
References
- Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, … Site—NDRI BCS (2017). Genetic effects on gene expression across human tissues. Nature, 550(7675), 204–213. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, … Geschwind DH (2015). The psychencode project. Nature Neuroscience, 18(12), 1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselmans BML, Jansen R, Ip HF, van Dongen J, Abdellaoui A, van de Weijer MP, … Bartels M (2019). Multivariate genome-wide analyses of the well-being spectrum. Nature Genetics, 51(3), 445–451. 10.1038/s41588-018-0320-8 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S, Rajaraman P, Jacobs KB, Wheeler WA, Melin BS, Hartge P, … Chatterjee N (2012). A Subset-Based Approach Improves Power and Interpretation for the Combined Analysis of Genetic Association Studies of Heterogeneous Traits. The American Journal of Human Genetics, 90(5), 821–835. 10.1016/j.ajhg.2012.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdeli TB, Genovese G, Georgakopoulos P, Meyers JL, Peterson RE, Iyegbe CO, … Kotov R (2019). Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Molecular Psychiatry, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady KT, Killeen T, & Jarrell P (1993). Depression in alcoholic schizophrenic patients. American Journal of Psychiatry, 150(8), 1255–1256. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, … Neale BM (2015). An atlas of genetic correlations across human diseases and traits. Nature Genetics, 47, 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, … Consortium, S. W. G. of the P. G. (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature Genetics, 47(3), 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Carniglia A, Keyes KM, Hasin DS, & Cerdá M (2019). Psychiatric comorbidities in alcohol use disorder. The Lancet Psychiatry, 6(12), 1068–1080. 10.1016/S2215-0366(19)30222-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T-K, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, … McIntosh AM (2017, July). Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112,117). Molecular Psychiatry. The Author(s). 10.1038/mp.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, S. W. G. of the P. G. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager PL, Ma Y, McCabe C, Xu J, Vardarajan BN, Felsky D, … Bennett DA (2018). A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Scientific Data, 5(1), 180142. 10.1038/sdata.2018.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, & Posthuma D (2015). MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Computational Biology, 11(4). 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, … Bækvad-Hansen M (2019). Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nature Genetics, 51(1), 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake RE, & Wallach MA (1989). Substance Abuse Among the Chronic Mentally Ill. Psychiatric Services, 40(10), 1041–1046. 10.1176/ps.40.10.1041 [DOI] [PubMed] [Google Scholar]
- Duke PJ, Pantelis C, & Barnes TRE (1994). South Westminster Schizophrenia Survey. Alcohol use and its relationship to symptoms, tardive dyskinesia and illness onset. British Journal of Psychiatry, 164(MAY), 630–636. 10.1192/bjp.164.5.630 [DOI] [PubMed] [Google Scholar]
- Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, … Consortium, T. B. (2018). Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nature Genetics, 50(4), 621–629. 10.1038/s41588-018-0081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA (1934). Statistical methods for research workers. Statistical Methods for Research Workers., (5th Ed). [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, … Sklar P (2016). Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nature Neuroscience. 10.1038/nn.4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum AS, Johnson EC, Polimanti R, Zhou H, Walters R, Consortium, S. U. D. W. G. of the P. G., … Agrawal A (2021). The Addiction Genetic Factor a(g): A Unitary Genetic Vulnerability Characterizes Substance Use Disorders and Their Associations with Common Correlates. MedRxiv, 2021.01.26.21250498. 10.1101/2021.01.26.21250498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, … Wigmore EM (2019). Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nature Neuroscience, 22(3), 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt GE, Large MM, Cleary M, Lai HMX, & Saunders JB (2018). Prevalence of comorbid substance use in schizophrenia spectrum disorders in community and clinical settings, 1990–2017: Systematic review and meta-analysis. Drug and Alcohol Dependence, 191, 234–258. [DOI] [PubMed] [Google Scholar]
- Johnson EC, Demontis D, Thorgeirsson TE, Walters RK, Polimanti R, Hatoum AS, … Agrawal A (2020). A large-scale genome-wide association study meta-analysis of cannabis use disorder. The Lancet Psychiatry. 10.1016/S2215-0366(20)30339-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Wang J-C, Farris SP, Liu Y, McClintick J, Gupta I, … Goate A (2019). Analysis of whole genome-transcriptomic organization in brain to identify genes associated with alcoholism. Translational Psychiatry, 9(1), 89. 10.1038/s41398-019-0384-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, … Gelernter J (2019). Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nature Communications, 10(1), 1499. 10.1038/s41467-019-09480-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M, Hill WD, Trampush JW, Yu J, Knowles E, Davies G, … Lencz T (2019). Pleiotropic Meta-Analysis of Cognition, Education, and Schizophrenia Differentiates Roles of Early Neurodevelopmental and Adult Synaptic Pathways. The American Journal of Human Genetics, 105(2), 334–350. 10.1016/J.AJHG.2019.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PH, Anttila V, Won H, Feng Y-CA, Rosenthal J, Zhu Z, … Smoller JW (2019). Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell, 179(7), 1469–1482.e11. 10.1016/j.cell.2019.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, & Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics, 27(12), 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, … Psychiatry HA-I (2019). Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nature Genetics, 51(2), 237–244. 10.1038/s41588-018-0307-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, … Moore HF (2013). The Genotype-Tissue Expression (GTEx) project. Nature Genetics, 45, 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Hu Y, Sun J, Cheng Y, Cheung K-H, & Zhao H (2015). A statistical framework to predict functional non-coding regions in the human genome through integrated analysis of annotation data. Scientific Reports, 5, 10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Li B, Ou D, Erlendsdottir M, Powles RL, Jiang T, … Zhao H (2017). A Powerful Approach to Estimating Annotation-Stratified Genetic Covariance via GWAS Summary Statistics. The American Journal of Human Genetics, 101(6), 939–964. 10.1016/j.ajhg.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Powles RL, Abdallah S, Ou D, Wang Q, Hu Y, … Mukherjee S (2017). Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease. PLoS Genetics, 13(7), e1006933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Powles RL, Wang Q, He BJ, & Zhao H (2016). Integrative tissue-specific functional annotations in the human genome provide novel insights on many complex traits and improve signal prioritization in genome wide association studies. PLoS Genetics, 12(4), e1005947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marees AT, Smit DJA, Abdellaoui A, Nivard MG, van den Brink W, Denys D, … Derks EM (2020). Genetic correlates of socio-economic status influence the pattern of shared heritability across mental health traits. MedRxiv, 2020.02.26.20028092. 10.1101/2020.02.26.20028092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marees AT, Smit DJA, Ong J-S, MacGregor S, An J, Denys D, … Derks EM (2019). Potential influence of socioeconomic status on genetic correlations between alcohol consumption measures and mental health. Psychological Medicine, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, … Lein ES (2014). Transcriptional landscape of the prenatal human brain. Nature, 508(7495), 199–206. 10.1038/nature13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, … Hamshere ML (2018). Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nature Genetics, 50(3), 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PF S, KS K, & MC N (2003). Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies. Archives of General Psychiatry, 60(12), 1187–1192. [DOI] [PubMed] [Google Scholar]
- Reginsson GW, Ingason A, Euesden J, Bjornsdottir G, Olafsson S, Sigurdsson E, … Stefansson K (2017). Polygenic risk scores for schizophrenia and bipolar disorder associate with addiction. Addiction Biology. 10.1111/adb.12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer DM, Ripke S, McQuillin A, Boocock J, Stahl EA, Pavlides JMW, … Kendler KS (2018). Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell, 173(7), 1705–1715.e16. 10.1016/j.cell.2018.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAMHSA. (2019). 2018 National Survey on Drug Use and Health (NSDUH).
- Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, 23andMe Research Team, Substance Use Disorder Working Group of the Psychiatric Genomics Consortium, … Clarke T-K (2018). Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in Two Population-Based Cohorts. American Journal of Psychiatry. 10.1176/appi.ajp.2018.18040369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, … Consortium, the B. D. W. G. of the P. G. (2019). Genome-wide association study identifies 30 loci associated with bipolar disorder. Nature Genetics, 51(5), 793–803. 10.1038/s41588-019-0397-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst B, Neale MC, & Kendler KS (2015). The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychological Medicine, 45(5), 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, … Team, 23andMe Research. (2018). Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nature Neuroscience, 21(12), 1656–1669. 10.1038/s41593-018-0275-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Stringer S, Frei O, Umićević Mirkov M, de Leeuw C, Polderman TJC, … Posthuma D (2019). A global overview of pleiotropy and genetic architecture in complex traits. Nature Genetics. 10.1038/s41588-019-0481-0 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, van Bochoven A, & Posthuma D (2017). Functional mapping and annotation of genetic associations with FUMA. Nature Communications, 8(1), 1826. 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, … Vos T (2013). Global burden of disease attributable to mental and substance use disorders: Findings from the Global Burden of Disease Study 2010. The Lancet, 382(9904), 1575–1586. 10.1016/S0140-6736(13)61611-6 [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, & Abecasis GR (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics, 26(17), 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Rentsch CT, Cheng Z, Kember RL, Nunez YZ, Sherva RM, … Program VAMV (2020). Association of OPRM1 Functional Coding Variant With Opioid Use Disorder: A Genome-Wide Association Study. JAMA Psychiatry, 77(10), 1072–1080. 10.1001/jamapsychiatry.2020.1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Sealock JM, Sanchez-Roige S, Clarke T-K, Levey DF, Cheng Z, … Gelernter J (2020). Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nature Neuroscience, 23(7), 809–818. 10.1038/s41593-020-0643-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, … Yang J (2016). Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nature Genetics, 48(5), 481–487. 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, … Wray NR (2018). Causal associations between risk factors and common diseases inferred from GWAS summary data. Nature Communications, 9(1), 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


