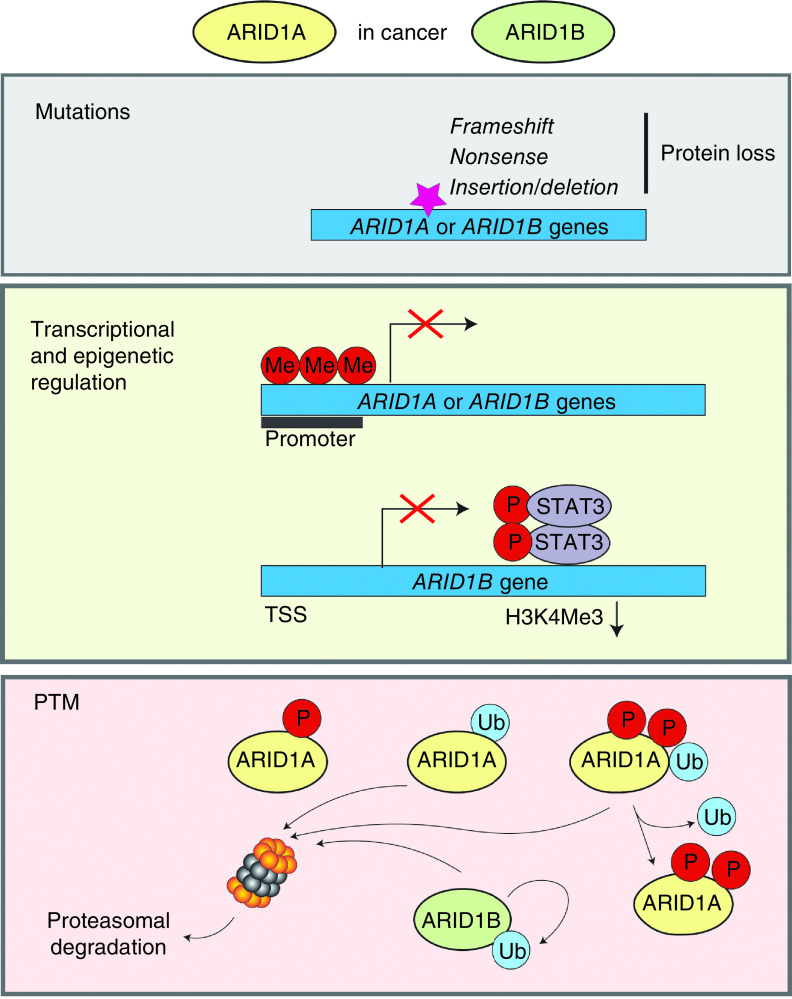
Figure 1. . Regulation of the ARID1-containing proteins in cancer.
The downregulation of ARID1 proteins in cancer is attributed to both mutational and non-mutational events. Most mutations in genes encoding ARID1 proteins include nonsense, frameshift and insertion/deletion mutations, and result in the protein loss. ARID1 genes are transcriptionally downregulated by their promoter methylation and repressive histone modifications. P-Stat3 transcriptionally represses ARID1B expression by decreasing H3K4Me3 histone modification. The levels of ARID1 proteins is controlled by ubiquitin-proteasome system. In gastric cancer, the DNA-damage repair signaling promotes rapid SCF β-TRCP-dependent ubiquitination and subsequent degradation of ARID1A. The phosphorylation of ARID1A at two serine sites by nuclear kinase ATM is required for its recognition by β-TRCP. In squamous cell carcinoma, ARID1A stability is regulated by E3 ligase TRIM32 and deubiquitinase USP11. ARID1B is not only a subunit of the BAF complexes but also a component of E3 ubiquitin ligase complex. ARID1B-harboring mutations in the BC box are less stable compared with wild-type ARID1B, and undergo autoubiquitination and proteasome-dependent degradation. In CCA-type ovarian cancer cell lines, p-Ser696-ARID1A levels is reduced compared with non-CCA cells possibly because of reduced phosphorylation or downregulation.
PTM: Post-translational modifications.