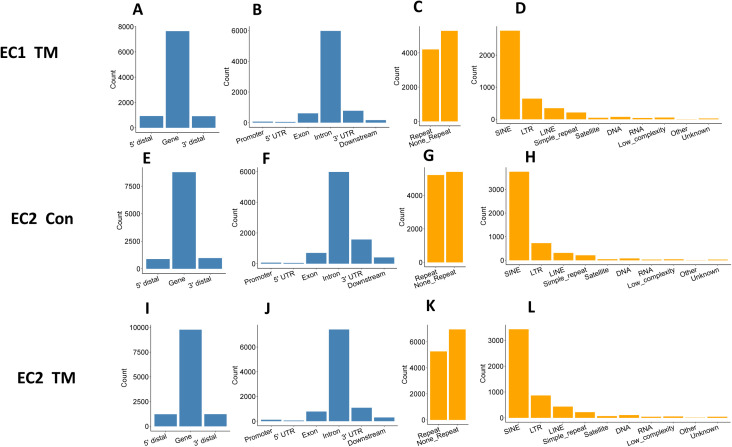
Figure S3. Distribution of RNA editing sites in endothelial cells.
Four independent RNA libraries were used for editing site identification and analysis, which were prepared from endothelial cells of two independent i-KO mice with and without tamoxifen (TM) induction. Results of EC1 control are shown in Fig 6. (A, B, C, D, E, F, G, H, I, J, K, L) Shown here are the results for EC1 treated with TM (EC1 TM, panel A, B, C, D), untreated EC2 (EC2 Con, panel E, F, G, H), and EC2 treated with TM (EC2 TM, panel I, J, K, L). (A, B, E, F, I, J) show that most editing sites fell into gene regions, especially intron and 3′ untranslated regions. (C, D, G, H, K, L) show that about half of the editing sites were in repetitive regions, most of them falling in short interspersed nuclear element, long terminal repeat, and long interspersed nuclear element.