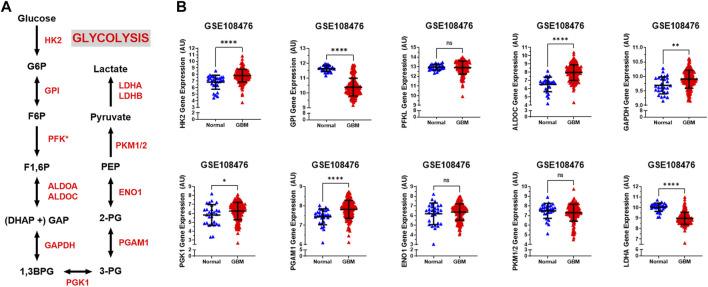
FIGURE 1.
Glycolytic genes are significantly overexpressed in GBM compared with normal tissue. (A) Schematic overview of glycolysis. Abbreviations of the enzymes are as follows: hexokinase 2 (HK2), glucose-6-phosphate isomerase (GPI), phosphofructokinase liver isoform (PFKL), aldolase A (ALDOA), glyceraldehyde 3 phosphate dehydrogenase (GAPDH), phosphoglycerate kinase 1 (PGK1), phosphoglycerate mutase 1 (PGAM), enolase 1 (ENO1), and pyruvate kinase M2 (PKM2), lactate dehydrogenase (LDH). Abbreviations of the metabolites are as follow: glucose-6-phosphate (G6P), fructose-6-phosphate (F6P), fructose 1,6-biphosphate (F1,6BP), glyceraldehyde 3-phosphate (GAP), and dihydroxyacetone phosphate (DHAP), 1,3-biphosphoglycerate (1,3BPG), glycerol-3-phosphate (3-PG), glycerol-2-phosphate (2-PG), phosphoenolpyruvate (PEP) (B) Relative gene expression of different glycolytic enzymes in the clinical data set GSE108476 consisting of GBM brain tissues (n= 174) and normal (n= 28) brain tissues. Gene expression data from the Repository for Molecular Brain Neplasia Data (REMBRANDT) via Affymetrix HG U133 v2.0 Plus. Significance calculated via Mann-Whitney Test. * indicates p < 0.05, ** indicates p < 0.001, **** indicates p < 0.0001, N/S indicates not significant.