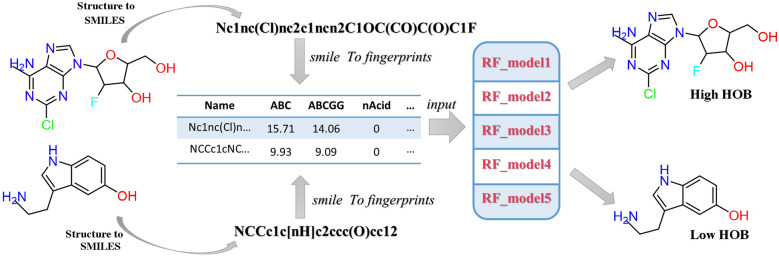
Graphical Abstract
Human oral bioavailability (HOB) is a key factor in determining the fate of new drugs in clinical trials. HOB is conventionally measured using expensive and time-consuming experimental tests. The use of computational models to evaluate HOB before the synthesis of new drugs will be beneficial to the drug development process. In this study, a total of 1588 drug molecules with HOB data were collected from the literature for the development of a classifying model that uses the consensus predictions of five random forest models. The consensus model shows excellent prediction accuracies on two independent test sets with two cutoffs of 20% and 50% for classification of molecules. The analysis of the importance of the input variables allowed the identification of the main molecular descriptors that affect the HOB class value. The model is available as a web server at www.icdrug.com/ICDrug/ADMET for quick assessment of oral bioavailability for small molecules. The results from this study provide an accurate and easy-to-use tool for screening of drug candidates based on HOB, which may be used to reduce the risk of failure in late stage of drug development.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13321-021-00580-6.
Keywords: Classification, Oral bioavailability, ADMET, Molecular descriptors, Prediction
Introduction
Poor pharmacokinetic properties, including absorption, distribution, metabolism, excretion, and toxicity (ADMET), are the key reasons of late-stage failures in drug development [1]. Therefore, ADMET assessments of candidate compounds during the early stages of drug discovery have become critical for improving the success rate of drug discovery and reducing the risk of late-stage attrition [2]. However, experimental testing of ADMET properties is time-consuming and costly. Thus, the accurate prediction of these properties is becoming increasingly important in drug discovery.
Among the ADMET properties, one of the most important pharmacokinetic characteristics of newly developed drugs is high oral bioavailability. Because oral administration is convenient and does not damage the skin or mucous membranes, 80% of the world’s drugs are administered orally [3]. Human oral bioavailability (HOB) is an important pharmacokinetic parameter that measures the amount of a drug that actually enters circulation within the body after ingestion. If intravenous administration is used, the human body can use the blood to deliver the drug to the site where it can exert pharmacological effects through the systemic circulation. Higher oral availability of the drug can reduce the amount of administration required to achieve the expected pharmacological effect because it can reduce the side effects and toxicity risks brought by the drug. On the other hand, poor oral bioavailability can lead to inefficiency of drugs and high inter-individual variability in the use of drugs, triggering some unpredictable drug reactions in the human body. In the actual drug development process, approximately 50% of candidate drugs fail due to low oral availability [4, 5]. Therefore, the level of oral availability is one of the key factors determining the success or failure of clinical trials of new drugs.
Experimental measurements of drug HOB are not only expensive, but also particularly time-consuming. Therefore, the development of a predictive model that can evaluate the HOB of a candidate compound before synthesis is of great help to drug discovery. Because the oral availability of a drug is affected by various biological, physical and chemical factors, such as the solubility of chemicals in the gastrointestinal tract, the permeability of the intestinal membrane, and the first pass metabolism of the intestine and liver, it is a very difficult and challenging task to develop accurate models to predict HOB. Nonetheless, a number of prediction models based on quantitative structure property relationships (QSPR) have been published [6, 7]. For example, Falcón-Cano et al. used 1448 molecules and obtained a consensus model with an accuracy of 78% [8]. Yang et al. used the random forest algorithm and 995 molecules to develop the admetSAR method with an accuracy of 69.7% [9]. On the basis of 995 data points, Kim et al. obtained 76% accuracy using the logistic classifier method [10].
In this study, we collected an HOB dataset composed of 1588 molecules and proposed a new model for HOB prediction based on machine learning. Using random forest (RF) [11–13] and two cutoffs of 20% and 50% for classifying molecules, we developed consensus models with a state-of-the-art accuracy on two independent test sets. Moreover, the importance of input molecular features to the prediction results was analyzed using the SHapley Additive exPlanation (SHAP) algorithm [14], revealing key molecular properties that affect HOB.
Materials and methods
Classification of positive and negative data
The performance of a classifier strongly depends on how the positive and negative samples are defined. However, there is still no consensus criterion to define positive and negative samples in HOB prediction. In previous studies, four cutoff values have been used: 20% (if HOB ≥ 20%, then the molecule belongs to the positive class; otherwise, it is a negative example) [15], 30% [16], 50% [17], and 80% [5] (Additional file 1: Table S1). We have therefor used two cutoffs 50% and 20% for labeling molecules in this study, which are used in more recent methods.
Dataset preparation
The HOB training and test datasets from Falcón-Cano et al. [8] were used in this study, which includes 1157 training and 290 test molecules. The 290 test molecules (test set 1) were selected by randomly selecting 20% of all molecules. Three molecules in the test set 1 had wrong values and were manually corrected according to the relevant literatures [18–21]. An additional test set of 27 molecules was collected from a number of publications [8] and was then combined with the HOB data from ChEMBL to form an additional test set of 141 molecules (Test set 2, Table 1). To ensure that molecules in test set 2 do not overlap with the molecules in training set and test set 1, 2D structures were first generated and used to remove duplicates followed by deduplication using molecular fingerprints. Neither test set 1 nor test set 2 was used during the training.
Table 1.
Information of the training and test datasets
| Cutoff | Data sets | Molecules | Positive | Negative |
|---|---|---|---|---|
| F = 50% | Training set | 1157 | 536 | 621 |
| Test set 1 | 290 | 169 | 121 | |
| Test set 2 | 141 | 90 | 51 | |
| F = 20% | Training set | 1142 | 859 | 283 |
| Test set 1 | 287 | 214 | 73 | |
| Test set 2 | 133 | 128 | 5 |
The labeling of molecules using the 50% cutoff was obtained directly from Falcón-Cano et al. For the 20% cutoff, some molecules cannot be classified due to inaccurate experimental values. These molecules were discarded, leaving training set with 1142 molecules, test set 1 with 287 molecules and test set 2 with 133 molecules (Table 1).
All molecules in the training and test sets were converted to 3D structures using the RDKit package. However, the calculated 3D fingerprints and descriptors were not used during model training due to null value or small variance for many of the molecules.
To evaluate the similarity between the training set and the test sets, we calculated the max Tanimoto coefficient similarity of each molecule in the test sets with all molecules in the training set. The similarity ranges from 0.1 to 1 in test set 1 and 0.2 to 1 in test set 2 (Fig. 1). The average similarity between the test set 1 and the training set is 0.655, and the average similarity between the test set 2 and the training set is 0.612.
Fig. 1.
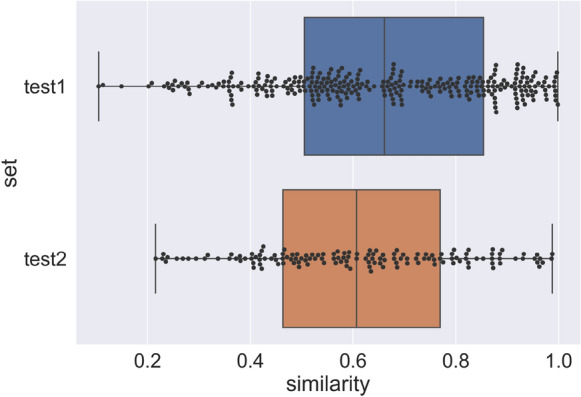
Maximum similarity of each molecule in the test sets to the molecules in the training set. The similarity was calculated using the Tanimoto coefficient similarity on the topological fingerprints of the molecules
Calculation of descriptors
We used Mordred [22] software to calculate 1614 molecular descriptors and fingerprints (Additional file 1: Table S2). Descriptors that had zero values or zero variance for all compounds were removed, reducing the total number of descriptors to 1143. All these 1143 features were used for training RF models.
Evaluation of models
The performance of the individual and consensus models was evaluated by analyzing the sensitivity (SE), specificity (SP), accuracy of prediction (ACC), Matthew’s correlation coefficient (MCC) [23] and F1_score.
where TP denotes true positive, FP is false positive, FN is false negative, and TN is true negative. In addition, the receiver operating characteristic (ROC) curve and the area under the curve (AUC) were also calculated.
We utilized the SHapley Additive exPlanation (SHAP) algorithm to explain the prediction model by providing consistent and locally accurate attribution values (SHAP values) for each feature within each prediction model [24]. The SHAP values evaluate the importance of the output resulting from the inclusion of a certain feature A for all combinations of features other than A.
Results
Performance of the models and comparison with previous methods
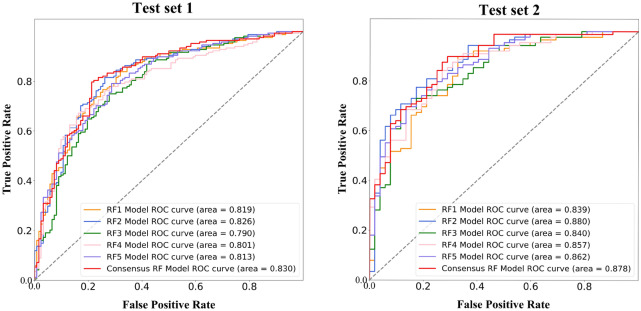
For the 50% cutoff, we first assembled a training set of 1157 molecules and two independent test sets with 290 and 146 molecules from the literature and public databases (see Materials and methods). The input features of the models include 1143 2D descriptors calculated with the Mordred package (see Materials and methods for details). Five individual RF models were first trained on the training set with fivefold cross-validation, using grid search and accuracy score (ACC) for hyperparameter optimization to obtain the best n_estimators and min_samples_leaf values while leaving the remaining hyperparameters set to default values (Table 2). Restricting the number of tunable hyper-parameters may reduce the risk of overfitting. The accuracy of the our model is 0.86–0.90 on the training set (Additional file 1: Tables S3, S4), and 0.74–0.77 on test set 1 (Table 3). The moderate decrease of accuracy on the test set suggests that our model has certain extend of overfitting but not severe. We then combined the five RF models to obtain a voting model. The final bioavailability class is the result of voting from each classification model with equal weight. Individual models can usually identify different aspects of the relationship between independent variables and dependent variables, and the relationships between the variables identified by those models may be different. In certain cases, the usage of a consensus model can greatly reduce the prediction error [25–27]. The accuracy of the five individual models ranges from 0.742 to 0.808 on the two test sets (Table 3). The consensus model shows improvement in accuracy on test set 1 and test set 2. The AUC values of the consensus model were 0.830 and 0.878 on the two test sets (Fig. 2). In addition, we merged the training set and test set 1, and used the same protocol to train five random forest models and obtained a consensus model from fivefold cross-validation. This whole process was repeated 50 times, and the average accuracy on test 2 was 0.826 with a standard deviation of 0.014, which was close to the accuracy of 0.823 when test set 1 is not included for training.
Table 2.
Optimized parameters of the RF models from fivefold cross-validation on the training set when the cutoff is 50%
| Parameters | Parameters meaning | Optimal value |
|---|---|---|
| n_estimators | The number of trees in the forest | 31 |
| min_samples_leaf | The minimum number of samples required to be at a leaf node | 6 |
Table 3.
Performance of the RF models on the two test sets when the cutoff is 50%
| Data set | Model | SE | SP | ACC | AUC | MCC | F1-score |
|---|---|---|---|---|---|---|---|
| Test set 1 | Model 1 | 0.779 | 0.732 | 0.752 | 0.819 | 0.505 | 0.774 |
| Model 2 | 0.746 | 0.786 | 0.769 | 0.826 | 0.529 | 0.798 | |
| Model 3 | 0.713 | 0.750 | 0.734 | 0.790 | 0.460 | 0.766 | |
| Model 4 | 0.745 | 0.732 | 0.738 | 0.801 | 0.473 | 0.764 | |
| Model 5 | 0.713 | 0.768 | 0.745 | 0.813 | 0.479 | 0.777 | |
| Consensus model | 0.787 | 0.797 | 0.793 | 0.830 | 0.580 | 0.817 | |
| Test set 2 | Model 1 | 0.673 | 0.899 | 0.816 | 0.839 | 0.596 | 0.860 |
| Model 2 | 0.673 | 0.876 | 0.801 | 0.880 | 0.565 | 0.848 | |
| Model 3 | 0.577 | 0.854 | 0.752 | 0.840 | 0.452 | 0.813 | |
| Model 4 | 0.692 | 0.865 | 0.801 | 0.857 | 0.568 | 0.846 | |
| Model 5 | 0.673 | 0.831 | 0.773 | 0.862 | 0.509 | 0.822 | |
| Consensus model | 0.692 | 0.899 | 0.823 | 0.872 | 0.612 | 0.865 |
Bold numbers refer to the maximum value (optimum value) obtained from the corresponding evaluation index
Fig. 2.
The AUCs of RF models on the two test sets when the cutoff is 50%
To further estimate the accuracy of the models, the accuracy of our model on test set 1 was compared with those of previously published HOB models (Table 4). It should be noted that we used the same training and test sets as Falcón-Cano et al., but the accuracies of other models are directly taken from the respective literature publications, which may use different data sets.
Table 4.
Comparison with other prediction models on test set 1 when the cutoff is 50%
| Model | Data set size | Method | ACC (test) | AUC (test) | Cut-off value |
|---|---|---|---|---|---|
| Current study | 1588 | RF | 0.793 | 0.830 | F = 50% |
| Falcón-Cano et al. [8] | 1448 | CART, MLP, NB, GBT, SVM | 0.783a | 0.800a | F = 50% |
| admetSAR [9] | 995 | RF | 0.697a | 0.752a |
logK(%F)b = 0 (F = 50%) |
| Kim et al. [10] | 995 | RF, SVM-consensus CTG | 0.76a | NA | F = 50% |
CART classification and regression trees; MLP multilayer perceptron; NB naive Bayes; GBT gradient boosted trees; SVM support vector machines
aTaken from respective references
b
Among these reported models, only admetSAR and ADMETlab provide an online prediction server that enables a direct comparison with our method based on the same test data. However, ADMETlab used different cutoffs of 30% and 20%. Therefore, we compared our method with admetSAR, which also uses F = 50% as the cut-off value. For a fair comparison, the molecules in the admetSAR training set were removed from the two test sets. Our model is more accurate than admetSAR in terms of SP, ACC and AUC (Table 5).
Table 5.
Comparison with admetSAR on test sets 1 and 2 when the cutoff is 50%
| Data set | Model | SE | SP | ACC | AUC |
|---|---|---|---|---|---|
| Test set 1a | Current Study | 0.824 | 0.819 | 0.821 | 0.862 |
| admetSAR | 0.784 | 0.777 | 0.780 | 0.831 | |
| Test set 2a | Current Study | 0.692 | 0.825 | 0.773 | 0.849 |
| admetSAR | 0.769 | 0.725 | 0.742 | 0.787 |
Bold numbers refer to the maximum value (optimum value) obtained from the corresponding evaluation index
aTest set 1 and 2 contain 168 and 66 molecules after removing the molecules used in the admetSAR training set
For the F = 20% cutoff, we used the same method to build a consensus model (Table 6). The accuracy of the five individual models ranges from 0.739 to 0.932 on the two test sets (Table 7). The consensus model shows improvement in accuracy on test set 1 and test set 2. The AUC values of the consensus model were 0.804 and 0.978 on the two test sets.
Table 6.
Optimized parameters of the RF models from fivefold cross-validation on the training set when the cutoff is 20%
| Parameters | Parameters meaning | Optimal value |
|---|---|---|
| n_estimators | The number of trees in the forest | 10 |
| min_samples_leaf | The minimum number of samples required to be at a leaf node | 6 |
Table 7.
Performance of the RF models on the two test sets when the cutoff is 20%
| Data set | Model | SE | SP | ACC | AUC | MCC | F1-score |
|---|---|---|---|---|---|---|---|
|
Test set 1 (287) |
Model 1 | 0.370 | 0.869 | 0.742 | 0.736 | 0.264 | 0.824 |
| Model 2 | 0.479 | 0.864 | 0.767 | 0.767 | 0.360 | 0.847 | |
| Model 3 | 0.452 | 0.893 | 0.780 | 0.771 | 0.379 | 0.858 | |
| Model 4 | 0.452 | 0.856 | 0.746 | 0.759 | 0.308 | 0.832 | |
| Model 5 | 0.493 | 0.855 | 0.763 | 0.789 | 0.359 | 0.833 | |
| Consensus model | 0.493 | 0.925 | 0.815 | 0.801 | 0.473 | 0.882 | |
|
Test set 2 (133) |
Model 1 | 0.8 | 0.891 | 0.887 | 0.973 | 0.384 | 0.938 |
| Model 2 | 1 | 0.859 | 0.865 | 0.978 | 0.432 | 0.924 | |
| Model 3 | 0.8 | 0.883 | 0.880 | 0.939 | 0.371 | 0.933 | |
| Model 4 | 1 | 0.914 | 0.917 | 0.955 | 0.534 | 0.955 | |
| Model 5 | 1 | 0.898 | 0.902 | 0.973 | 0.499 | 0.946 | |
| Consensus model | 1 | 0.906 | 0.910 | 0.981 | 0.516 | 0.951 |
Bold numbers refer to the maximum value (optimum value) obtained from the corresponding evaluation index
The performance of our model is compared with that of ADMETlab [16] on the two test sets (Table 8). Our model showed lower ACC and AUC on test set 1 but better performance on test set 2. Because test set 1 comes from an earlier data set that was published before ADMETlab [8, 10, 28–30], and has overlapping molecules with the training sets of several previous methods, it is likely that some molecules in test set 1 may be included in the training of ADMETlab (We were not able to obtain the training set of ADMETlab for removing of redundancy). On the other hand, the HOB data in test set 2 does not overlap with any published training sets and may server as a more objective testing of the two models.
Table 8.
Comparison with ADMETlab on test sets 1 and 2 when the cutoff is 20%
| Data set | Model | SE | SP | ACC | AUC |
|---|---|---|---|---|---|
| Test set 1 | Current study | 0.493 | 0.925 | 0.815 | 0.801 |
| ADMETlab | 0.904 | 0.855 | 0.868 | 0.947 | |
| Test set 2 | Current study | 1 | 0.906 | 0.910 | 0.981 |
| ADMETlab | 0.8 | 0.844 | 0.842 | 0.902 |
Bold numbers refer to the maximum value (optimum value) obtained from the corresponding evaluation index
Diversity distribution of HOB data
In this study, 1157, 290 and 141 molecules that have human oral availability data were collected for model construction. To examine the diversity of these molecules and provide a possible way to access the applicability of our model, we carried out principal component analysis (PCA) of these molecules. The 1143 fingerprints and descriptors mentioned above for model training were used to generate PCA for all compounds. We selected the two most important components to create a chemical space for characterizing training set, test set 1, and test set 2 (Fig. 3). The results suggest that the chemical space of the two test sets is roughly within the space of the training set, therefore it is sensible to use the prediction model trained by the training set to predict the HOB values for the test sets. In addition, after removing an outlier in test set 1 (circled points in Fig. 3) that are outside the PCA space of the training set, the accuracy remains unchanged on test set 2 with 50% and 20% cutoff. Moreover, the PCA analysis can be used to determine the applicability of our model to new molecules. When the projection of a new molecule is within the range of the training molecules, it is considered as “inside” the application domain, indicating a more reliable prediction.
Fig. 3.
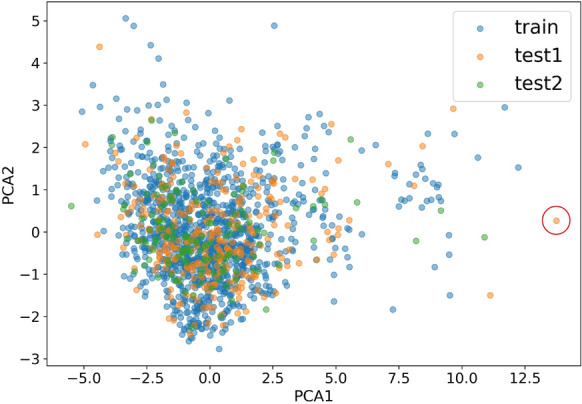
The chemical space of the training set (blue), the test set 1 (orange) and test set 2(green) using PCA factorization
Diversity evaluation of base learners
Diversity is very important in combining base learners. The Q-value approach is one way to measure the diversity of two classifiers [31]. It ranges between − 1 and 1, and is 0 if two classifiers are independent. The larger the Q value, the smaller the difference between the predictions of two classifiers. We used Q-value to measure the difference between the decision trees in each model. The average Q-value for individual trees in the five random forest models when the cutoff is 50% was 0.207, 0.233, 0.27, 0.241, and 0.267. When the cutoff is 20%, the Q-values were 0.235, 0.269, 0.288, 0.293, and 0.275, which suggest that these trees have high diversity.
Importance of the input features
The analysis of important descriptors and fingerprints for prediction provides more information to fully understand these models. To this end, we used the SHapley Additive exPlanation (SHAP) algorithm to calculate the importance score of the input descriptor and fingerprints [14]. SHAP (SHapley Additive exPlanations) is a game theory method used to explain the output of a machine learning model. It uses the classical Shapley value from game theory and its related extension to link the optimal credit allocation with local explanation.
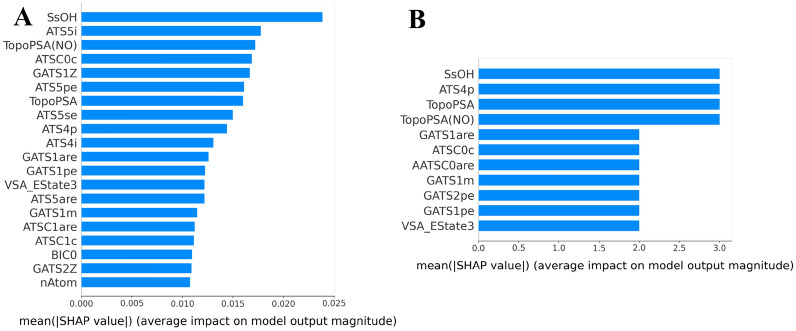
We used the same method to analyze the contribution of each descriptor in the RF models when the cutoff is 50%, and the top 20 most important variables that contributed to the model were obtained through importance matrix plots (Additional file 1: Figs. S1–S5). The importance matrix plot for the consensus method was calculated by averaging the value in each model (Fig. 4A), which depicts the importance of each input feature in the development of the final predictive model. SsOH (number of all atoms) which is an atom type e-state descriptor contributes the most to predictive power, followed by the topological structure descriptor ATS5i and polar surface area descriptor TopoPSA (NO). In addition, we also counted the number of appearances of the features in the five individual models (Fig. 4B). SsOH, ATS4p and TopoPSA(NO) appear three times among the 20 most important descriptors of the five models. In the two methods of quantifying feature importance, the specific information of the top 20 features is sorted in Table 9.
Fig. 4.
A Importance matrix plot of the consensus model when the cutoff is 50%. B A statistical graph of the number of occurrences of the top 20 features that affect all models when the cutoff is 50%
Table 9.
Description of the important features in the consensus model when the cutoff is 50%
| Descriptor category | Feature name | |shap value| | Description |
|---|---|---|---|
| Estate | SsOH | 0.0239 | Sum of sOH |
| TopoPSA | TopoPSA(NO) | 0.0178 | Topological polar surface area (use only nitrogen and oxygen) |
| Autocorrelation | ATSC0c | 0.0172 | Centered Moreau–Broto autocorrelation of lag 0 weighted by Gasteiger charge |
| Autocorrelation | GATS1Z | 0.0169 | Moreau–Broto autocorrelation of lag 1 weighted by atomic number |
| Autocorrelation | ATS5pe | 0.0167 | Moreau–Broto autocorrelation of lag 5 weighted by Pauling EN |
| TopoPSA | TopoPSA | 0.0161 | Topological polar surface area |
| Autocorrelation | ATS5se | 0.0160 | Moreau–Broto autocorrelation of lag 5 weighted by Sanderson EN |
| Autocorrelation | ATS4p | 0.0150 | Moreau–Broto autocorrelation of lag 4 weighted by polarizability |
| Autocorrelation | ATS4i | 0.0144 | Moreau–Broto autocorrelation of lag 4 weighted by ionization potential |
| Autocorrelation | GATS1are | 0.0131 | Geary coefficient of lag 1 weighted by Allred–Rocow EN |
| Autocorrelation | GATS1pe | 0.0126 | Geary coefficient of lag 1 weighted by Pauling EN |
| MoeType | VSA_EState3 | 0.0122 | VSA EState Descriptor 3 (5.00 ≤ x < 5.41) |
| Autocorrelation | ATS5are | 0.0122 | Moreau–Broto autocorrelation of lag 5 weighted by Allred–Rocow EN |
| Autocorrelation | GATS1m | 0.0122 | Geary coefficient of lag 1 weighted by mass |
| Autocorrelation | ATSC1are | 0.0115 | Centered Moreau–Broto autocorrelation of lag 1 weighted by Allred–Rocow EN |
| Autocorrelation | ATSC1c | 0.0112 | Centered Moreau–Broto autocorrelation of lag 1 weighted by Gasteiger charge |
| InformationContent | BIC0 | 0.0111 | 0-ordered bonding information content |
| Autocorrelation | GATS2Z | 0.0110 | Geary coefficient of lag 2 weighted by atomic number |
| AtomCount | nAtom | 0.0109 | Molecular ID on O atoms |
The SHAP dependence plot is further used to understand how a single feature affects the output of the prediction models, where the color bar indicates the actual values of the features, and the SHAP values are plotted on the x-axis. The dependence plot of the consensus model when the cutoff is 50% was obtained by averaging the contributions of the individual models (Fig. 5, Additional file 1: Figs. S6–10). For each feature, a dot is created for each molecule, and positive SHAP values for a specific feature represent an increase in the predicted HOB value. For example, TopoPSA (NO) (topological polar surface area, using only nitrogen and oxygen) is an important feature that is overall negatively correlated with HOB (Fig. 5), i.e., a higher TopoPSA(NO) value will reduce the predicted HOB value. This is consistent with the finding that reducing the polar surface area increases the permeation rate of a molecule [32]. Similarly, the TopoPSA descriptor which calculates the entire polar surface area is also negatively correlated with the HOB value.
Fig. 5.
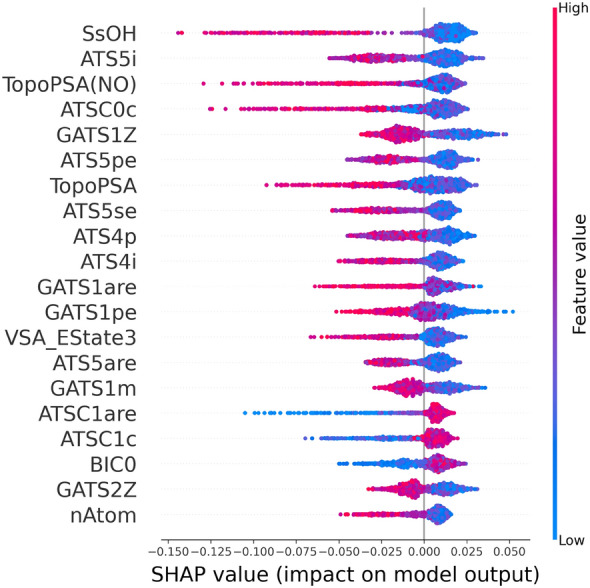
SHAP dependence plot of the top 20 features of the consensus model when the cutoff is 50%
In addition, SsOH which is the total number of OH bonds also significantly affects the prediction of HOB. The blue dots are mainly concentrated in the area where the SHAP is greater than 0, therefore a small SsOH value will increase the HOB value. Decreasing the number of OH groups will increase the hydrophobicity and membrane absorption of a molecule, therefore leading to higher HOB. This is in line with the Lipinski's ‘Rule-of-Five’ [33]: if the number of hydrogen bond donors exceeds 5, the absorption or permeability may be poor [34].
It is believed that the charge state of molecules exerts a key influence on the perception of biomolecules (including membranes, enzymes and transporters) [35]. Several features that have great influence in the consensus model, such as RPCG in RF1, ATSC0c in RF2 and RNCG in RF4 are charge related descriptors.
Using the same method, we also analyzed the model obtained with the 20% cutoff, and obtained importance matrix plots (Additional file 1: Figs. S11–S15, S21) and the dependence plots (Additional file 1: Figs. S16–S20, S22) of the individual and the consensus models. The important features from the models trained with the two cutoffs are overall consistent, e.g., TopoPSA (NO), SsOH and Autocorrelation also have a significant impact on the F = 20% consensus model as that on the F = 50% model.
Discussion and conclusions
On the basis of a comprehensive data sets collected in this study, accurate RF models for prediction of HOB were developed. Moreover, we also analyzed the importance of the features, and found that the number of OH bonds and polar surface area of a molecule are negatively correlated with the HOB value, which is consistent with molecular characteristics that affect the oral drug bioavailability. The model is available as a web server at www.icdrug.com/ICDrug/ADMET, which provides researchers with an accurate tool to quickly and automatically predict the HOB of new molecules without any machine learning or statistical modeling knowledge.
Supplementary Information
Additional file 1: Table S1. Cut-off values used in different studies and the number of positive and negative samples in each study. Table S2. List of descriptors calculated using Mordred. Table S3. The performance of the consensus model on the training set and each fold in the fivefold cross validation when the cutoff is 50%. Table S4. The performance of the consensus model on the training set and each fold in fivefold cross validation when the cutoff is 20%. Figure S1. The importance matrix plot for the RF model 1 when the cutoff is 50%. Figure S2. The importance matrix plot for the RF model 2 when the cutoff is 50%. Figure S3. The importance matrix plot for the RF model 3 when the cutoff is 50%. Figure S4. The importance matrix plot for the RF model 4 when the cutoff is 50%. Figure S5. The importance matrix plot for the RF model 5 when the cutoff is 50%. Figure S6. SHAP dependence plot of the top 20 features of the RF model 1 when the cutoff is 50%. Figure S7. SHAP dependence plot of the top 20 features of the RF model 2 when the cutoff is 50%. Figure S8. SHAP dependence plot of the top 20 features of the RF model 3 when the cutoff is 50%. Figure S9. SHAP dependence plot of the top 20 features of the RF model 4 when the cutoff is 50%. Figure S10. SHAP dependence plot of the top 20 features of the RF model 5 when the cutoff is 50%. Figure S11. The importance matrix plot for the RF model 1 when the cutoff is 20%. Figure S12. The importance matrix plot for the RF model 2 when the cutoff is 20%. Figure S13. The importance matrix plot for the RF model 3 when the cutoff is 20%. Figure S14. The importance matrix plot for the RF model 4 when the cutoff is 20%. Figure S15. The importance matrix plot for the RF model 5 when the cutoff is 20%. Figure S16. SHAP dependence plot of the top 20 features of the RF model 1 when the cutoff is 20%. Figure S17. SHAP dependence plot of the top 20 features of the RF model 2 when the cutoff is 20%. Figure S18. SHAP dependence plot of the top 20 features of the RF model 3 when the cutoff is 20%. Figure S19. SHAP dependence plot of the top 20 features of the RF model 4 when the cutoff is 20%. Figure S20. SHAP dependence plot of the top 20 features of the RF model 5 when the cutoff is 20%. Figure S21. (A) Importance matrix plot of the consensus model when the cutoff is 20%. (B) A statistical graph of the number of occurrences of the top 20 features that affect all models when the cutoff is 20%. Figure S22. SHAP dependence plot of the top 20 features of the consensus model when the cutoff is 20%.
Acknowledgements
The computer time was provided by the ECNU Multifunctional Platform for Innovation001.
Abbreviations
- HOB
Human oral bioavailability
- ADMET
Absorption, distribution, metabolism, excretion, and toxicity
- QSPR
Quantitative structure property relationships
- SHAP
The SHapley Additive exPlanation
- SE
Sensitivity
- SP
Specificity
- ACC
Accuracy of prediction
- MCC
Matthew’s correlation coefficient
- ROC
The receiver operating characteristic
- AUC
The receiver operating characteristic curve and the area under the curve
- RF
Random forest
- CART
Classification and regression trees
- MLP
Multilayer perceptron
- NB
Naive Bayes
- GBT
Gradient boosted trees
- SVM
Support vector machines
- PCA
Principal component analysis
Authors' contributions
YQ and JZ designed the research plan. MW, XZ, XP, and BW performed the computational analysis. MW and YQ wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key R&D Program of China (Grant No. 2016YFA0501700), the National Natural Science Foundation of China (22033001, 21933010), and the Natural Science Foundation of Shanghai (19ZR1473600).
Availability of data and materials
The training and test data sets and the trained models are available at https://github.com/whymin/HOB and the web server at www.icdrug.com/ICDrug/ADMET. Other data are available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yifei Qi, Email: yfqi@fudan.edu.cn.
John Z. H. Zhang, Email: John.zhang@nyu.edu
References
- 1.Waring MJ, Arrowsmith J, Leach AR, et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat Rev Drug Discov. 2015;14(7):475–486. doi: 10.1038/nrd4609. [DOI] [PubMed] [Google Scholar]
- 2.van de Waterbeemd H, Gifford E. ADMET in silico modelling: towards prediction paradise? Nat Rev Drug Discov. 2003;2(3):192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 3.Morishita M, Peppas NA. Advances in oral drug delivery: improved bioavailability of poorly absorbed drugs by tissue and cellular optimization. Preface. Adv Drug Deliv Rev. 2012;64(6):479. doi: 10.1016/j.addr.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy T. Managing the drug discovery/development interface. Drug Discov Today. 1997;2(10):436–444. doi: 10.1016/S1359-6446(97)01099-4. [DOI] [Google Scholar]
- 5.Ahmed SS, Ramakrishnan V. Systems biological approach of molecular descriptors connectivity: optimal descriptors for oral bioavailability prediction. PLoS ONE. 2012;7(7):e40654. doi: 10.1371/journal.pone.0040654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J, Wang J, Yu H, et al. Recent developments of in silico predictions of oral bioavailability. Comb Chem High Throughput Screen. 2011;14(5):362–374. doi: 10.2174/138620711795508368. [DOI] [PubMed] [Google Scholar]
- 7.Xiong G, Wu Z, Yi J, et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falcon-Cano G, Molina C, Cabrera-Perez MA. ADME prediction with KNIME: development and validation of a publicly available workflow for the prediction of human oral bioavailability. J Chem Inf Model. 2020;60(6):2660–2667. doi: 10.1021/acs.jcim.0c00019. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Lou C, Sun L, et al. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35(6):1067–1069. doi: 10.1093/bioinformatics/bty707. [DOI] [PubMed] [Google Scholar]
- 10.Kim MT, Sedykh A, Chakravarti SK, et al. Critical evaluation of human oral bioavailability for pharmaceutical drugs by using various cheminformatics approaches. Pharm Res. 2014;31(4):1002–1014. doi: 10.1007/s11095-013-1222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Svetnik V, Liaw A, Tong C, et al. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43(6):1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 12.Sheridan RP. Using random forest to model the domain applicability of another random forest model. J Chem Inf Model. 2013;53(11):2837–2850. doi: 10.1021/ci400482e. [DOI] [PubMed] [Google Scholar]
- 13.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 14.Lundberg SM, Lee SI (2017) A unified approach to interpreting model predictions. In: Advances in neural information processing systems, pp 4765–4774
- 15.Ma CY, Yang SY, Zhang H, et al. Prediction models of human plasma protein binding rate and oral bioavailability derived by using GA-CG-SVM method. J Pharm Biomed Anal. 2008;47(4–5):677–682. doi: 10.1016/j.jpba.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Dong J, Wang NN, Yao ZJ, et al. ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J Cheminform. 2018;10(1):29. doi: 10.1186/s13321-018-0283-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olivares-Morales A, Hatley OJ, Turner D, et al. The use of ROC analysis for the qualitative prediction of human oral bioavailability from animal data. Pharm Res. 2014;31(3):720–730. doi: 10.1007/s11095-013-1193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Rashidy R. Estimation of the systemic bioavailability of timolol in man. Biopharm Drug Dispos. 1981;2(2):197–202. doi: 10.1002/bdd.2510020213. [DOI] [PubMed] [Google Scholar]
- 19.Korte JM, Kaila T, Saari KM. Systemic bioavailability and cardiopulmonary effects of 0.5% timolol eyedrops. Graefes Arch Clin Exp Ophthalmol. 2002;240(6):430–5. doi: 10.1007/s00417-002-0462-2. [DOI] [PubMed] [Google Scholar]
- 20.Johansson SA, Knutsson M, Leonsson-Zachrisson M, et al. Effect of food intake on the pharmacodynamics of tenapanor: a phase 1 study. Clin Pharmacol Drug Dev. 2017;6(5):457–465. doi: 10.1002/cpdd.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.FDA Approved Drug Products: Lysteda (tranexamic acid) tablets for oral use. https://link.springer.com/content/pdf/10.1007%2F3-540-44938-8_31.pdf
- 22.Moriwaki H, Tian YS, Kawashita N, et al. Mordred: a molecular descriptor calculator. J Cheminform. 2018;10(1):4. doi: 10.1186/s13321-018-0258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh KP, Basant N, Gupta S. Support vector machines in water quality management. Anal Chim Acta. 2011;703(2):152–162. doi: 10.1016/j.aca.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 24.Strumbelj E, Kononenko I. An efficient explanation of individual classifications using game theory. J Mach Learn Res. 2010;11(1532–4435):1–18. [Google Scholar]
- 25.Lei T, Li Y, Song Y, et al. ADMET evaluation in drug discovery: 15. Accurate prediction of rat oral acute toxicity using relevance vector machine and consensus modeling. J Cheminform. 2016;8:6. doi: 10.1186/s13321-016-0117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu H, Martin TM, Ye L, et al. Quantitative structure-activity relationship modeling of rat acute toxicity by oral exposure. Chem Res Toxicol. 2009;22(12):1913–1921. doi: 10.1021/tx900189p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei B, Li J, Yao X. A novel strategy of structural similarity based consensus modeling. Mol Inform. 2013;32(7):599–608. doi: 10.1002/minf.201200170. [DOI] [PubMed] [Google Scholar]
- 28.Tian S, Li Y, Wang J, et al. ADME evaluation in drug discovery. 9. Prediction of oral bioavailability in humans based on molecular properties and structural fingerprints. Mol Pharm. 2011;8(3):841–51. doi: 10.1021/mp100444g. [DOI] [PubMed] [Google Scholar]
- 29.Varma MVS, Obach RS, Rotter C, et al. Physicochemical space for optimum oral bioavailability: contribution of human intestinal absorption and first-pass elimination. J Med Chem. 2010;53(3):1098–1108. doi: 10.1021/jm901371v. [DOI] [PubMed] [Google Scholar]
- 30.Dörwald FZ. Lead optimization for medicinal chemists: pharmacokinetic properties of functional groups and organic compounds. Weinheim, Germany: Wiley-VCH; 2012. [Google Scholar]
- 31.Kuncheva LI, Whitaker CJ. Measures of diversity in classifier ensembles and their relationship with the ensemble accuracy. Mach Learn. 2003;51(2):181–207. doi: 10.1023/A:1022859003006. [DOI] [Google Scholar]
- 32.Veber DF, Johnson SR, Cheng HY, et al. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 33.Lipinski CA, Lombardo F, Dominy BW, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 1997;23(1–3):3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- 34.Hou T, Wang J, Zhang W, et al. ADME evaluation in drug discovery. 6. Can oral bioavailability in humans be effectively predicted by simple molecular property-based rules? J Chem Inf Model. 2007;47(2):460–3. doi: 10.1021/ci6003515. [DOI] [PubMed] [Google Scholar]
- 35.Martin YC. A bioavailability score. J Med Chem. 2005;48(9):3164–3170. doi: 10.1021/jm0492002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Cut-off values used in different studies and the number of positive and negative samples in each study. Table S2. List of descriptors calculated using Mordred. Table S3. The performance of the consensus model on the training set and each fold in the fivefold cross validation when the cutoff is 50%. Table S4. The performance of the consensus model on the training set and each fold in fivefold cross validation when the cutoff is 20%. Figure S1. The importance matrix plot for the RF model 1 when the cutoff is 50%. Figure S2. The importance matrix plot for the RF model 2 when the cutoff is 50%. Figure S3. The importance matrix plot for the RF model 3 when the cutoff is 50%. Figure S4. The importance matrix plot for the RF model 4 when the cutoff is 50%. Figure S5. The importance matrix plot for the RF model 5 when the cutoff is 50%. Figure S6. SHAP dependence plot of the top 20 features of the RF model 1 when the cutoff is 50%. Figure S7. SHAP dependence plot of the top 20 features of the RF model 2 when the cutoff is 50%. Figure S8. SHAP dependence plot of the top 20 features of the RF model 3 when the cutoff is 50%. Figure S9. SHAP dependence plot of the top 20 features of the RF model 4 when the cutoff is 50%. Figure S10. SHAP dependence plot of the top 20 features of the RF model 5 when the cutoff is 50%. Figure S11. The importance matrix plot for the RF model 1 when the cutoff is 20%. Figure S12. The importance matrix plot for the RF model 2 when the cutoff is 20%. Figure S13. The importance matrix plot for the RF model 3 when the cutoff is 20%. Figure S14. The importance matrix plot for the RF model 4 when the cutoff is 20%. Figure S15. The importance matrix plot for the RF model 5 when the cutoff is 20%. Figure S16. SHAP dependence plot of the top 20 features of the RF model 1 when the cutoff is 20%. Figure S17. SHAP dependence plot of the top 20 features of the RF model 2 when the cutoff is 20%. Figure S18. SHAP dependence plot of the top 20 features of the RF model 3 when the cutoff is 20%. Figure S19. SHAP dependence plot of the top 20 features of the RF model 4 when the cutoff is 20%. Figure S20. SHAP dependence plot of the top 20 features of the RF model 5 when the cutoff is 20%. Figure S21. (A) Importance matrix plot of the consensus model when the cutoff is 20%. (B) A statistical graph of the number of occurrences of the top 20 features that affect all models when the cutoff is 20%. Figure S22. SHAP dependence plot of the top 20 features of the consensus model when the cutoff is 20%.
Data Availability Statement
The training and test data sets and the trained models are available at https://github.com/whymin/HOB and the web server at www.icdrug.com/ICDrug/ADMET. Other data are available from the corresponding author on reasonable request.