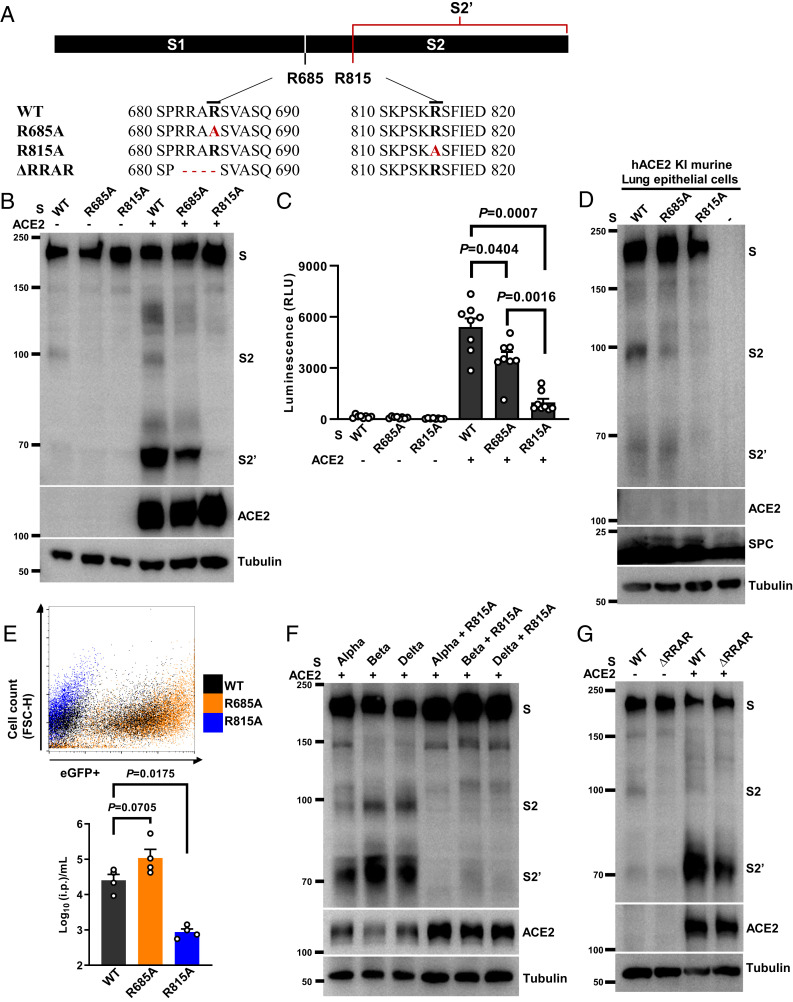
Fig. 4.
Spike arginine 815 is required for S2′ cleavage and syncytia formation. (A) Schematics and amino acid sequences depicting the relative positions of S1/S2 and S2′ cleavage sites along the spike protein; spike R685A and R815A substitutions and ΔRRAR are highlighted in red. (B) Immunoblots showing the full-length S, S2, S2′, and ACE2 collected from HEK293T cells expressing S-WT, S-R685A, or S-R815A cocultured with HEK293T cells without or with ACE2 expression for 16 h. Blots are representative of five independent experiments. (C) Luciferase activity (RLU) detected from S-WT, S-R685A, or S-R815A induced syncytia lysates. Data are shown as individual points with mean ± SEM from eight independent experiments. (D) Immunoblots showing the full-length S, S2, S2′, ACE2, SPC, and tubulin collected from HEK293T cells expressing S-WT, S-R685A, or S-R815A mixed with murine lung epithelial cells isolated from hACE2 KI mice. Blots are representative of two individual experiments. (E) Representative dot plot from FACS showing percentage of eGFP-positive HEK293T-ACE2 cells infected by SARS-CoV-2 PPs prepared using S-WT (Gray), S-R685A (Orange), or S-R815A (Blue) for 48 h. Numbers of i.p. per milliliter were calculated from infected HEK293T-ACE2 cells; data summarized from four individual repeats were presented as mean ± SEM. (F) Immunoblots showing S, S2, and S2′ generation in HEK293T cells transiently expressing spike Alpha, Beta, and Delta variants containing R815A mutations upon cocultured with HEK293T-ACE2 cells for 16 h. Blots are representative of three individual experiments. (G) Immunoblots showing the full-length S, S2, S2′, ACE2, and tubulin from HEK293T cells expressing WT orΔRRAR spike mutants mixed with HEK293T cells without or with ACE2 expression for 16 h. Blots are representative of three individual repeats. P values were obtained by one-way ANOVA with Sidak’s post hoc test and are indicated in C.