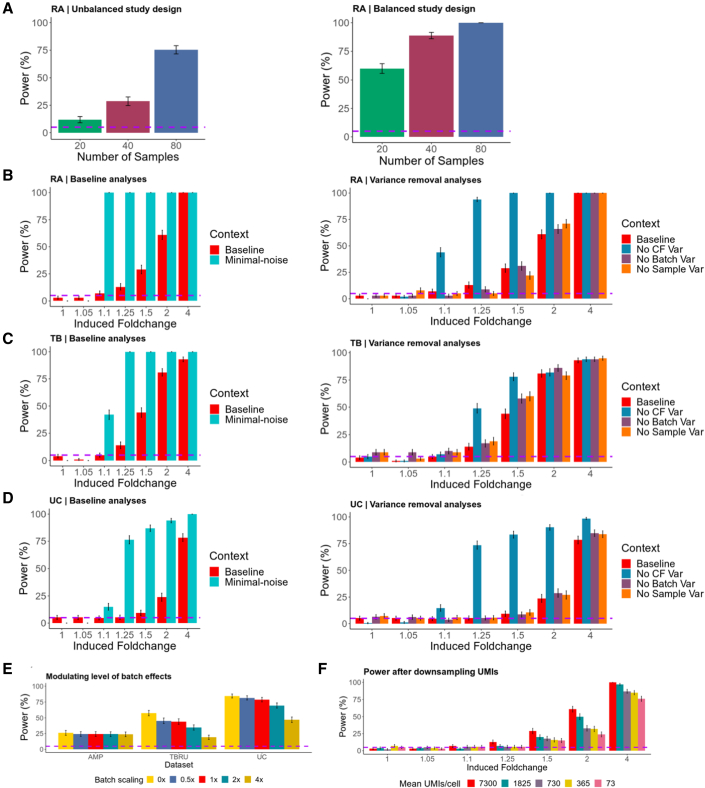
Figure 5.
scPOST estimates power from multiple study designs
Each bar represents n = 500 simulations. Error bars represent 95% binomial proportion confidence intervals; dotted horizontal purple lines are set at 5%.
(A) Results from increasing the size of a study.
(B–D) Left, Baseline/minimal-noise analyses for the RA, TB, and UC datasets. Baseline context features realistic levels of cluster frequency (CF) variation and gene expression variation (bscale = 1, sscale = 1, cfscale = 1). Minimal-noise removes these sources of variation (e.g., bscale = 0). Right, analyses in which only one source of variation (CF, batch, or sample) is removed.
(E) Batch scaling simulations: 0× indicates no batch variation, whereas 4× indicates 4 times the level of estimated realistic batch variation. Induced fold changes of 1.5 in RA and TB settings and fold change of 4 in UC.
(F) Power calculations derived from downsampling the RA UMI data (red = baseline).