FIGURE 6.
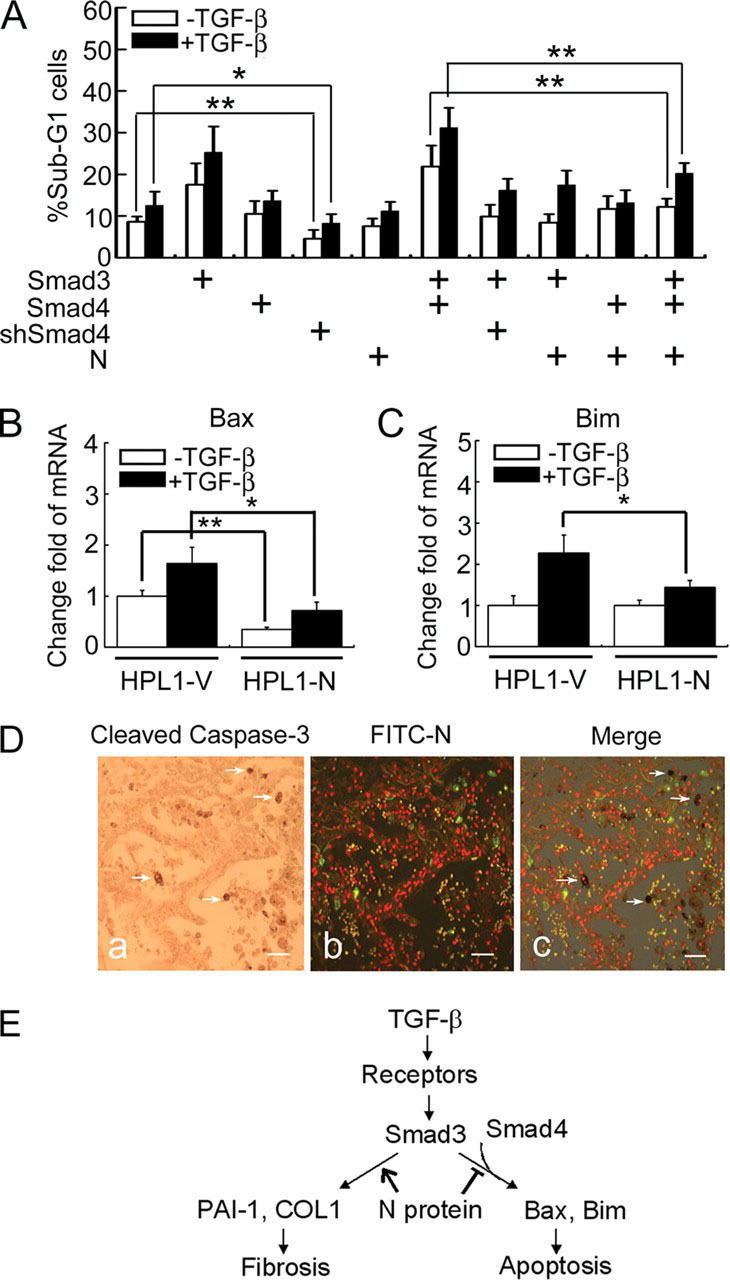
N protein impairs TGF-β-induced HPL1 cell apoptosis.A, N protein inhibits TGF-β-induced HPL1 cell apoptosis. HPL1 cells were co-transfected with GFP plasmid (0.1 μg), pCS2-Myc-Smad3 (1 μg), pCS2-FLAG-Smad4 (1 μg), pSUPER-siRNA against Smad4 (0.5 μg), and pcDNA3.1-N (1 μg) as indicated. Those GFP-positive cells were selected by FACS for DNA contents analysis. The percentage of sub-G1 cells in total GFP-positive cells was accounted for. Each experiment was repeated in triplicate, and the data represent the mean ± S.D. of three independent experiments. B and C, the mRNA levels of endogenous Bim and Bax in HPL1-V and HPL1-N were analyzed by real-time PCR. GAPDH served as a loading control. The asterisks indicate a statistically significant difference between HPL1-V and HPL1-N cells (*, p < 0.05; **, p < 0.01). D, N protein-positive cells are not co-localized with active caspase-3-positive apoptotic cells in SASR patient's lung. Apoptotic cells were detected by anti-cleaved caspase-3 antibody and developed by diaminobenzidine tetrahydrochloride (a). SARS-CoV-infected cells were detected by fluorescein isothiocyanate-labeled anti-N protein antibody and the image was taken with a fluorescence microscopy. The virus-infected cells were green (b). White arrows indicate the apoptotic cells in c. Scale bar:50 μm. E, a working model depicts the role of SARS-CoV N protein in modulating TGF-β/Smad3-induced fibrosis and apoptosis. The asterisks indicate a statistically significant difference (*, p < 0.05; **, p < 0.01).
