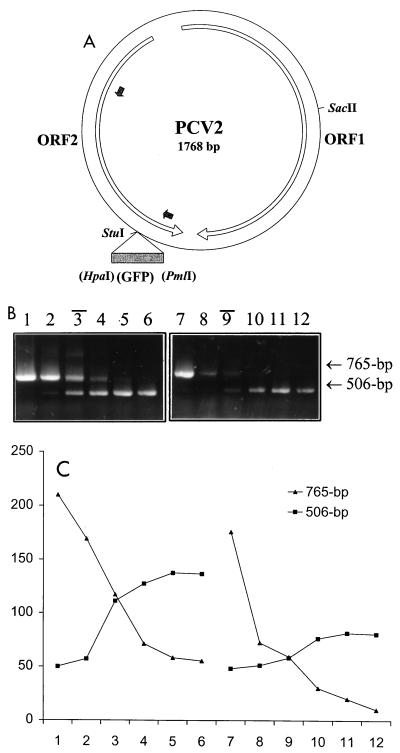
FIG. 1.
Construction of wild-type and competitor plasmids and validation of cPCR. (A) Diagrammatic representation of PCV2 wild-type and competitor constructs. The whole genome (1,768 nucleotides) of PCV2 was cloned with a SacII restriction enzyme into the pBKSII(+) vector. The largest two open reading frames (ORFs) in the PCV2 genome are indicated by open arrows. Two small filled arrows point out the primer binding sites. A portion of the GFP coding sequence as the insert for the construction of the competitor plasmid is demonstrated by a filled box. (B and C) Quantification of a known amount of PCV2 wild-type DNA. (B) cPCR assay performed with two series of 10-fold dilutions of competitor DNA (lanes 1 to 6, 100 ng, 10 ng, 1 ng, 100 pg, 10 pg, and 1 pg, respectively; lanes 7 to 12, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, and 1 fg, respectively) to compete against two fixed amount of PCV2 wild-type DNA per reaction (lanes 1 to 6, 1 ng; lanes 7 to 12, 1 pg). Black bars above the lane numbers indicate the equivalent point. (C) The densities of DNA bands were measured with SPOT DENSO software by using an AlphaImager 2200 instrument, and values representing the average density per area unit were plotted. The numbers on the x axis correspond to the respective lane in panel B. The value determined for the wild-type DNA was in excellent agreement with the actual input amount.