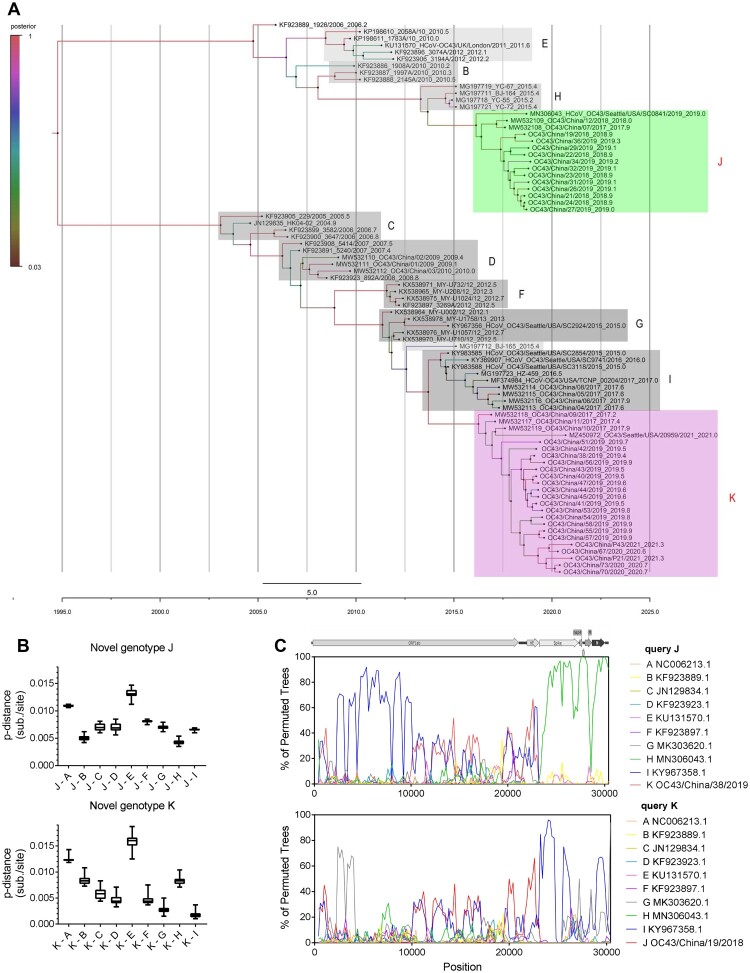
Figure 1.
Analysis of time-resolved phylogeny, pairwise genetic distances and genetic recombination of two novel genotypes of HCoV-OC43. (A) Beast software was used to conduct estimated divergence time analysis of novel genotypes of HCoV-OC43 based on spike gene. A HKY nucleotide substitution model with estimated base frequencies and a lognormal relaxed clock model were used for evolution analysis. The Markov chain Monte Carlo (MCMC) analysis was performed with 1 × 109 generations with every 10,000 generations were sampled. The sampling year, strain name and accession number are shown on labels. Colour on branch represented the posterior probability corresponding to the legend. Two novel genotypes are indicated in green (J) and pink (K), respectively. (B) Estimation of pairwise genetic distances of novel genotypes J and K to previous genotypes based on the full-length genome sequences, respectively. (C) Genetic recombinant analyses of HCoV-OC43 novel genotypes J and K.