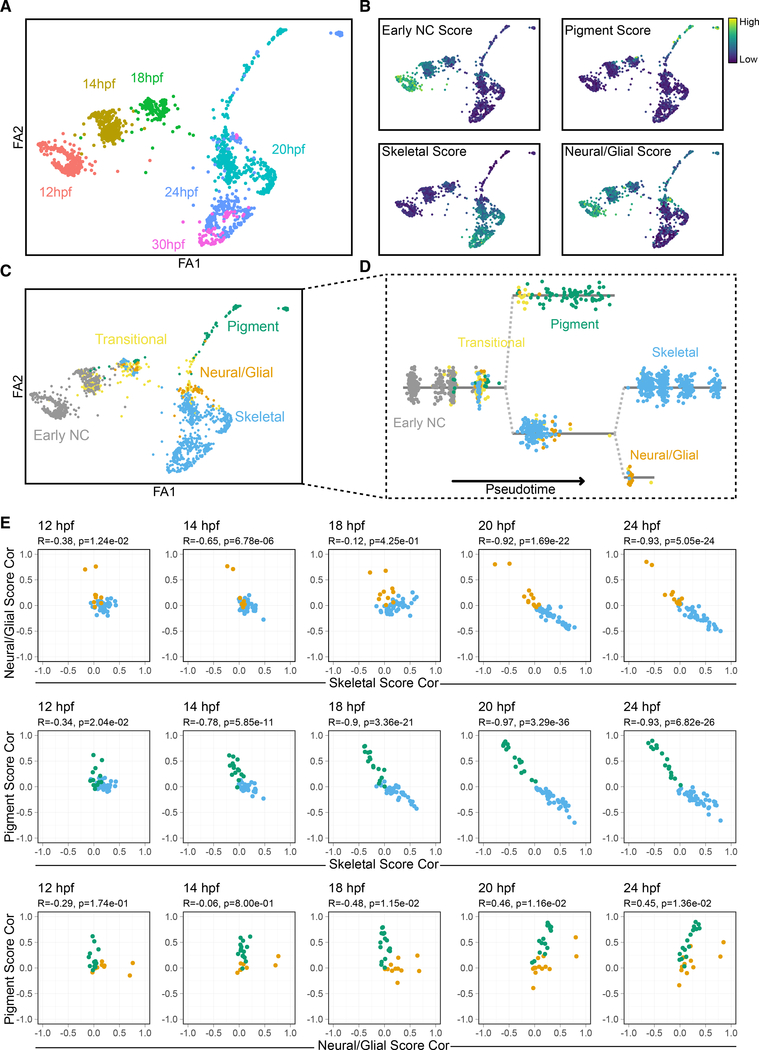
Figure 3. Pseudotemporal analysis reveals emergence of NC lineages during migration.
(A) ForceAtlas (FA) embedding of single-cell timeline initialized using PAGA graph with time point identities of individual cells labeled and color coded.
(B) Module scores, color code distinct from (A), for early NC, pigment, skeletal, and neural and glial lineages overlaid on FA embedding.
(C) CellAssign probabilistic cell type labels computed using module score genes overlaid on FA embedding. Color code is distinct from (A) or (B).
(D) Pseudotime calculated using PAGA graph. Cells are arranged according to pseudotime values along the x axis, with pseudotime scaled within each branch. Branch points are split on the y axis and connected by dashed lines. Color code corresponds to (C).
(E) Scatterplots showing the increased specificity of marker genes to lineages across developmental time. Each point represents a gene contributing to a lineage score (color code from C) with the axes showing its Pearson correlation to the total score for pigment, skeletal, or neural and glial across all cells in each time point.