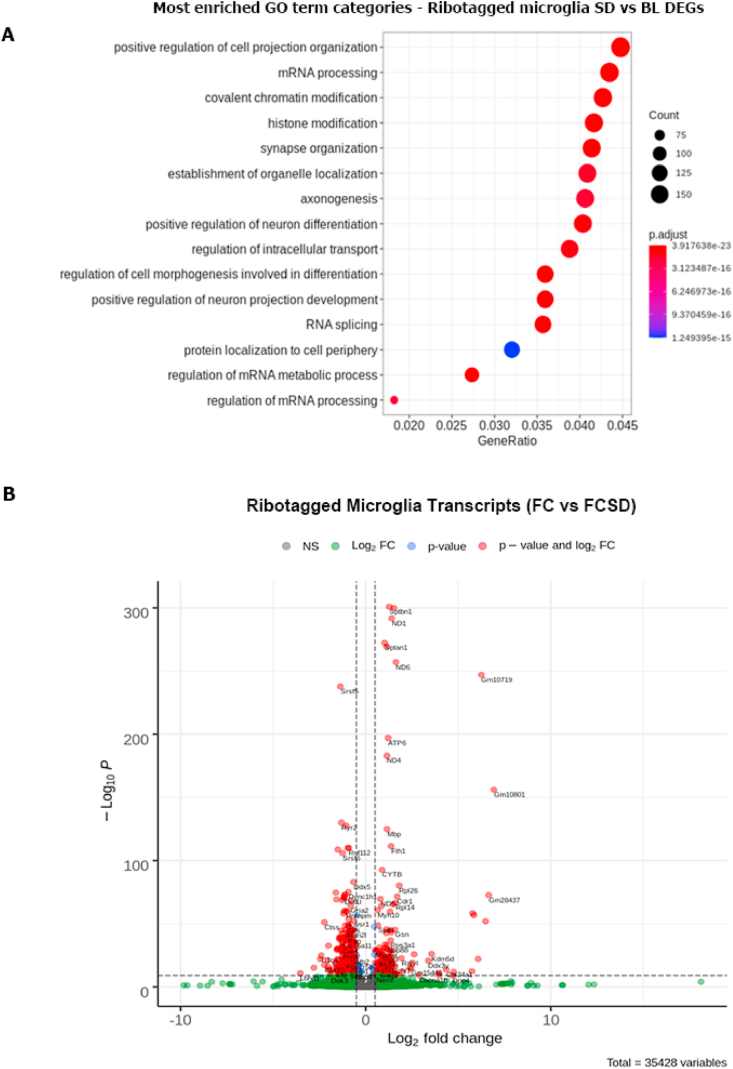
Fig. 3.
Sleep deprivation modifies the microglial transcriptome to activate synaptic-associated genes (A), RNA-seq analysis comparing mice at ZT = 6 following fear conditioning with and without SD. Mice were either sacrificed 6 h after FC 6 h of fear conditioning, at ZT = 6, with or without SD from ZT = 0 to ZT = 6. GO term analysis of a submitted list of ∼4000 differentially expressed genes to Python's clusterProfiler package which utilizes PANTHER's GO system. This analysis indicates relative enrichment for terms compared to a random sampling; the color of each circle represents the result of a relative enrichment analysis within each category compared to a random sample of genes, GeneRatio represents total proportion of sample, and size of circle indicates absolute number of genes in category. n = 4 for each condition. For detailed analysis on RNAseq processing, see methods. (B), Volcano plot of all genes identified in sample and analyzed for significance by DEseq2 created using EnhancedVolcano package. Cutoffs for coloring are at a -log10 value of 10 and at a 2 fold expression change. A selection of the top 50 most significantly differentially expressed genes were selected for labeling on the chart. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)