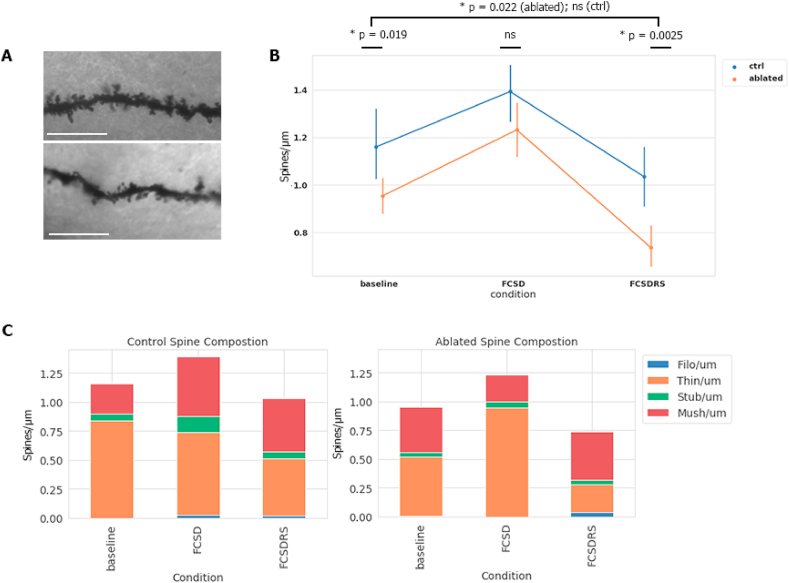
Fig. 4.
Microglial ablation leads to altered spine density dynamics (A), Representative image of Golgi staining in FCSD condition for control (top) and ablated (bottom). Only in-focus 10 μm stretches were measured. Scale bar = 10 μm. (B), Total spines with error values for each condition. Baseline = no intervention, ZT = 0; FCSD = fear conditioning plus 6 h of SD, ZT = 6; FCSDRS = fear conditioning plus 6 h of SD and 18 h subsequent recovery sleep, where samples were taken at ZT = 0. Different cohorts of mice used for each condition. Golgi stained 120 μm brain slices were selected and viewed at 100x magnification. Random visible 10 μm stretches of CA3 dendrite were chosen and imaged for downstream analysis. All comparisons between conditions and microglia status are statistically significant except for those shown as not-significant (ctrl BL vs FCSD; p = 0.010. ctrl FCSD vs FCSDRS; p < 0.001. ctrl BL vs ctrl FCSDRS; ns. abl BL vs FCSD; p = 0.0015. abl FCSD vs abl FCSDRS; p < 0.001. abl BL vs FCSDRS; p = 0.022). Statistical analysis performed using an unpaired parametric two-tailed student's T test between relevant conditions. (C), Spine density of each of spine type as categorized by the Reconstruct software, showing the same population of spines as (B) split by type. After the Reconstruct program was provided stretches of spines, it quantified and categorized the relative composition of each morphological spine type. Categories ‘thin’ and ‘long thin’ spines were combined for this analysis. Left; control spines in each condition, right; spines in microglia ablated mice. Data expressed as mean. n = 3 or 4 mice with 30-60 dendritic stretches. Related Fig. S4 displays this data in FC and SD contexts individually, and Table S2 provides statistical testing results between numerous spine type and context comparisons.