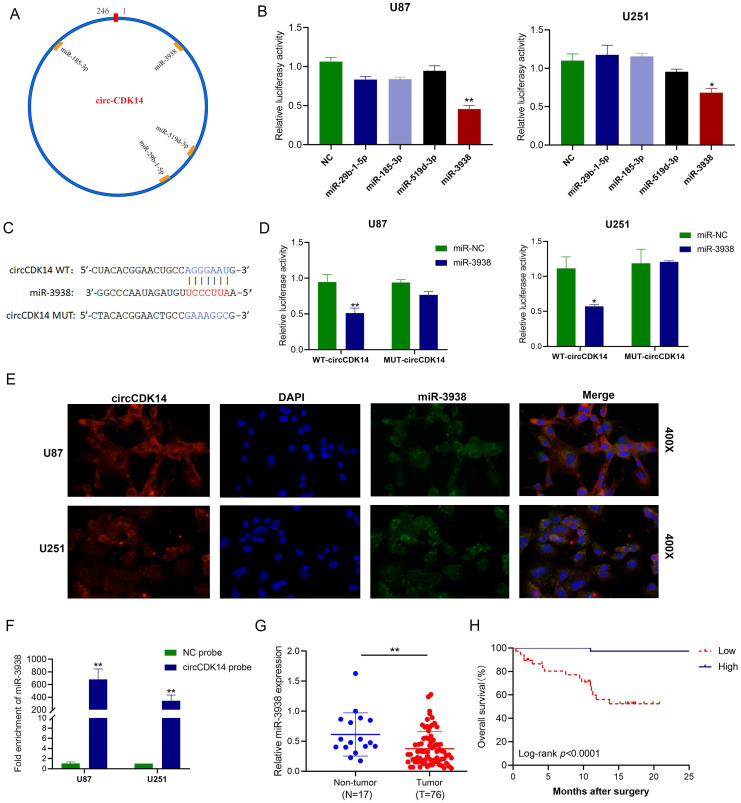
Figure 3.
circCDK14 acts as sponge for miR-3938 in glioma. (A) Illustrations of the estimated docking sites of miRNA candidates to circCDK14. (B) Luciferase activity of pmirGLO-circCDK14-WT in cells after co-incorporation with miRNAs mimics. (C) Estimated docking sites of miR-3938 with circCDK14 was predicted. circCDK14 WT, circCDK14 wild-type luciferase reporter; circCDK14 MUT, circCDK14 mutantluciferase reporter. (D) U251 and U87 cells were co-transfected with pmirGLO -circCDK14-WT or pmirGLO -circCDK14-MUT and miR‐3938 mimics or negative control. Luciferase activity measured with luciferase reporter assays. (E) Colocalization between circCDK14 and miR-3938 were observed by RNA in situ hybridization (FISH) in U87 and U251 cells. DAPI staining was used for the nuclei. CircCDK14 probe was Cy3-labeled, and miR-3938 probe was FAM-labeled. (F) RNA pull-down assay was conducted to show the relative miR-3938 level in U87 and U251 cells lysates pulled down by circCDK14. (G) The expression of miR-3938 in 76 glioma and 17 non-tumor brain tissues were detected by qRT-PCR. (H) The overall survival curve, based on miR-3938 levels, was plotted with Kaplan-Meier methods and analyzed by rank test. All experiments repeated three times and data averaged and expressed as mean±SD.*p< 0.05; **p< 0.01.