Figure 1. ccRCC cells harbor deregulated cholesterol metabolism.
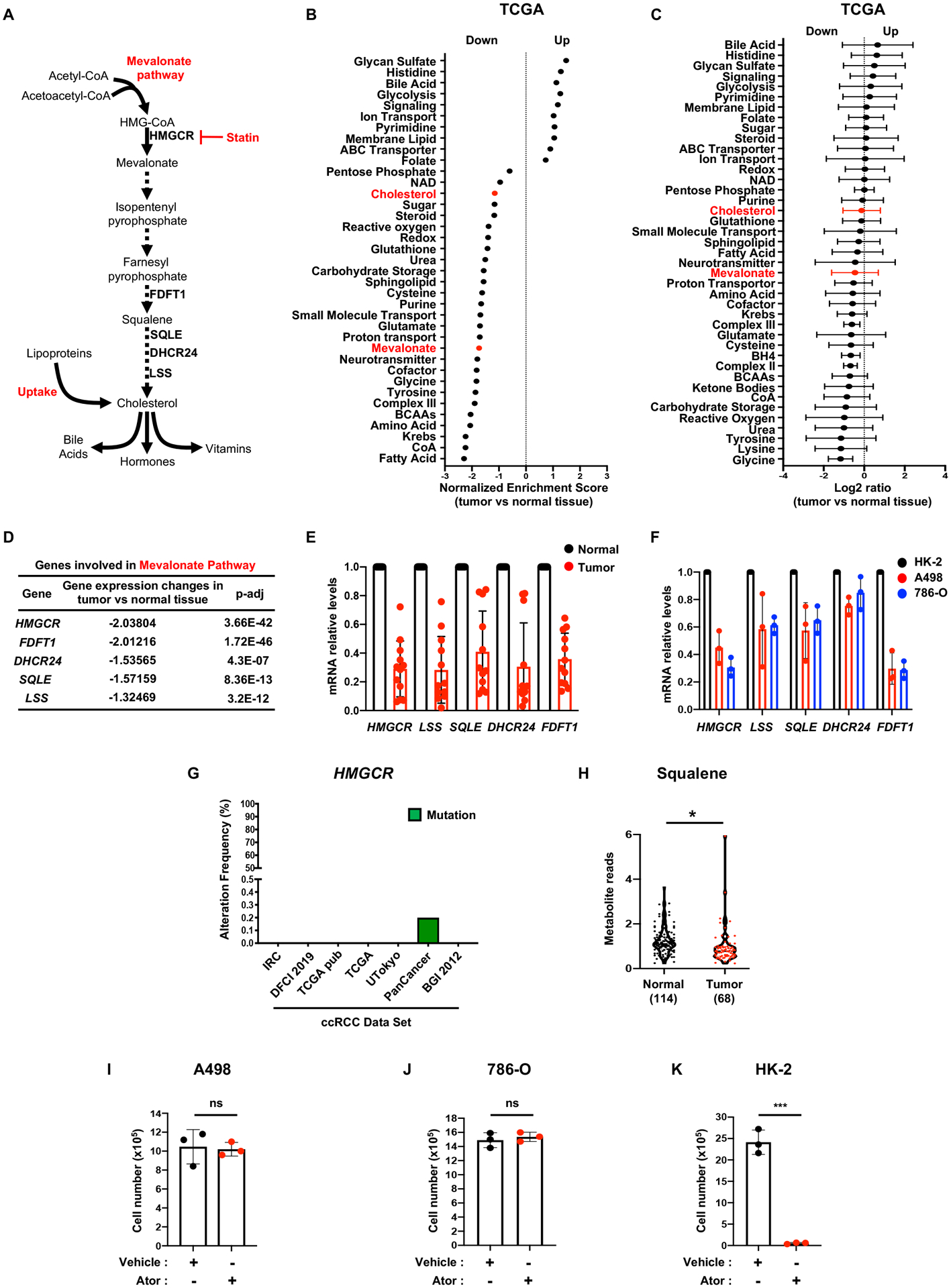
A, Simplified schematic of the mevalonate pathway. Mammalian cells maintain cholesterol homeostasis through direct synthesis, which can be inhibited by statins, or import from the extracellular environment.
B, Gene set enrichment analysis (GSEA) of RNAseq data provided by the TCGA KIRC project indicating that genes belonging to the “cholesterol” and “mevalonate” pathways have lower expression in ccRCC tumors compared to normal kidney tissue. Generated metabolic gene sets were ranked based on normalized enrichment score changes in ccRCC compared to normal tissue.
C, Metabolic gene set analysis of RNAseq data provided by the TCGA KIRC project. 538 ccRCC tumor and 72 adjacent normal tissues were included. 2,752 genes encoding all human metabolic enzymes and transporters were classified according to KEGG. Generated metabolic gene sets were ranked based on their log2 median fold expression changes in ccRCC compared to normal tissue.
D, TCGA dataset analysis shows that expression of genes involved in the “mevalonate pathway” is significantly downregulated in ccRCC tumors vs. normal tissue. HMGCR, 3-hydroxy-3-methylglutaryl-coenzyme A reductase; FDFT1, squalene synthase; DHCR24, delta 24-sterol reductase; SQLE, squalene monooxygenase; LSS, lanosterol synthase.
E, Real-time qPCR analysis performed on 12 tumor tissues and their normal counterparts, indicating mevalonate pathway gene expression is decreased in tumors compared to normal tissues.
F, Real-time qPCR analysis performed on immortalized proximal tubular renal epithelial cells (HK-2) and two ccRCC cell lines, A498 and 786-O, evaluating expression of HMGCR, LSS, SQLE, DHCR24 and FDFT1.
G, Alteration frequency of HMGCR gene in several kidney cancer genomic datasets using cBio Cancer genomic portal. IRC, Nat Genet 2012; DFCI, Science 2019; TCGA pub, firehose legacy; TCGA, Nature 2013; Utokyo, Nat Genet 2013, TCGA PanCancer Atlas; BGI, Nat Genet 2012.
H, Metabolomics analysis of squalene in 114 normal kidney tissues and 68 ccRCC tumors.
I, J and K, A498, 786-O and HK-2 cell proliferation assays showing insensitivity of ccRCC cell lines to 72h of atorvastatin (ATOR) treatment (5μM). (All experiments were performed in at least triplicates and statistical analysis was applied with *=P<0.05, **=P<0.01, ***=<0.001, n.s=non-significant).
