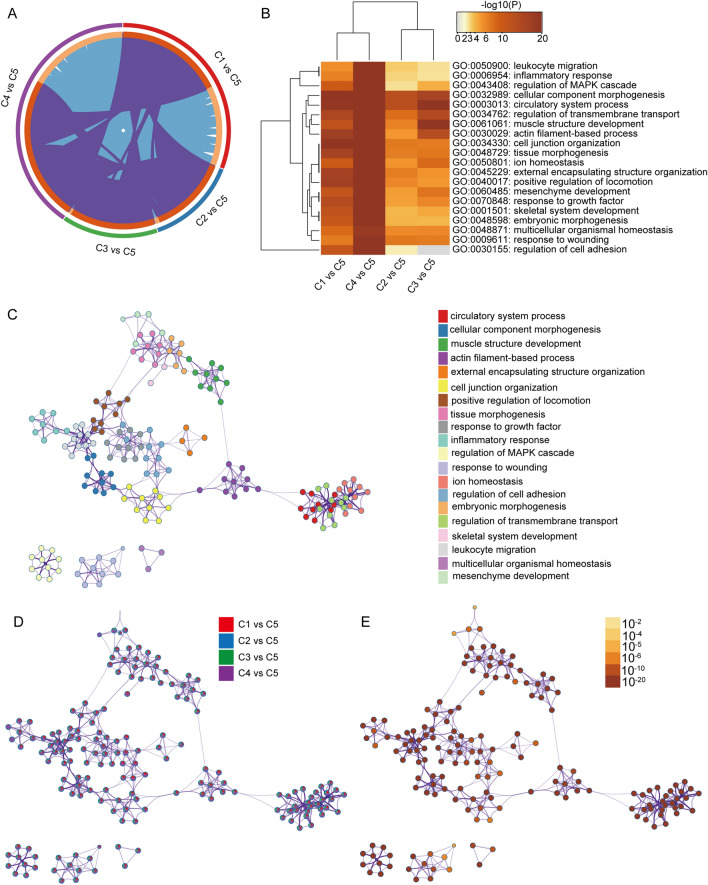
Figure 3.
Meta-enrichment analysis summary for PFNs lists in 4 comparison cohorts. (A). The overlap of differentially expressed genes in the four selected groups is shown in the circus plot explored using Metascape. (B). Each box indicates whether the gene in each gene list (column, C1 vs C5, C4 vs C5, C2 vs C5, and C3 vs C5) enriched in each selected top 20 GO term. The darker the color, the more significance the P-value. R package heatmap is used in this process, and two-way hierarchical clustering is used for clustering. (C). The network of enriched terms is colored by cluster-ID, and nodes that share the same cluster are typically close to each other. (D). Network enrichment terms and genes are colored by the database, where the terms containing more genes tends to have more significates. (E). Coloring by the degree of enrichment, with darker colors indicating a greater number of genes enriched to that pathway or biological process class.