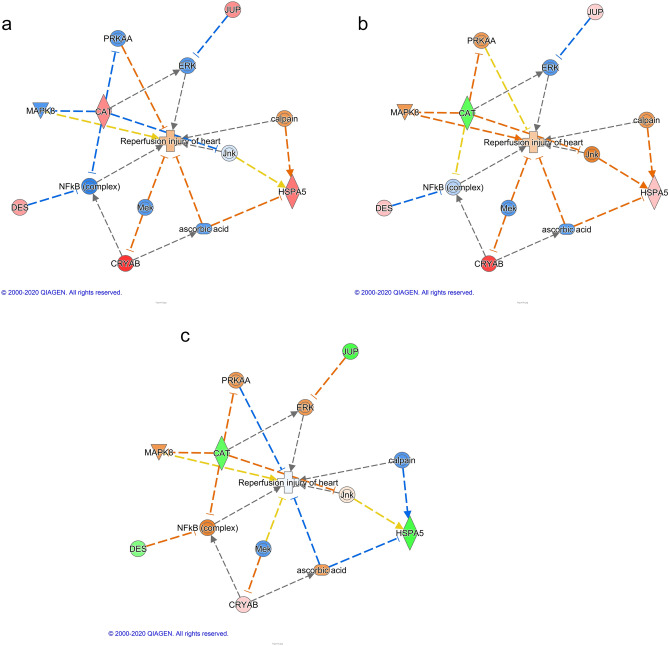
Figure 7.
Analysis of proteomic data with Ingenuity Pathway Analysis (IPA) in WT and CPNS1 KO (knockout) mitochondria. (a) Predicted effects using WT:IR/TC data in the proteins-IR network. (b) Predicted effects using KO:IR/TC data in the proteins-IR network. (c) Predicted effects using IR: KO/WT data in the proteins-IR network. The figure was generated in Qiagen Ingenuity Pathway Analysis (IPA) program using several differentially expressed proteins (pink or red for upregulated protein, green for downregulated proteins) and their relationships to reperfusion injury of heart. The relationships and several mediating proteins, complexes or endogenous chemicals were added using the Pathway Explorer tool in IPA, and the predicted effects (orange for activation, blue for inhibition, yellow indicates the finding is inconsistent with the state of downstream molecule, and grey for no prediction) were made by the Molecule Activity Predictor (MAP) of IPA. Relationships: 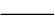 Direct interaction.
Direct interaction.  Indirect interaction.
Indirect interaction. 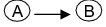 activation, causation, expression, localization, membership, modification, phosphorylation, regulation of binding, transcription.
activation, causation, expression, localization, membership, modification, phosphorylation, regulation of binding, transcription.  inhibition, ubiquitination.
inhibition, ubiquitination.