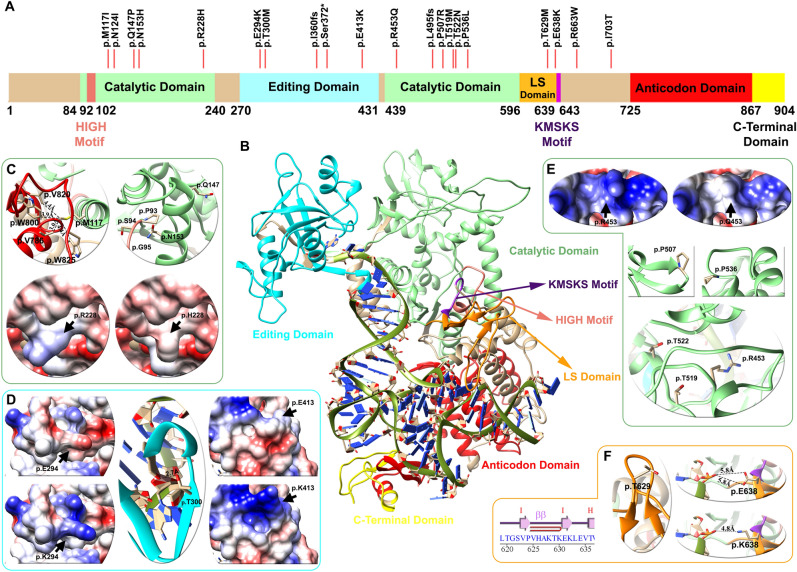
Figure 2.
Domain architecture, mapping of variants and structural analysis of the LeuRS protein (LARS2 gene). (A) Linear representation of the LeuRS protein with its domains and motifs: HIGH motif (pink), catalytic domain (light green), editing domain (cyan), LS domain (orange), KMSKS motif (purple), anticodon domain (red), and C-terminal domain (yellow). Red lines depict the location of the p.Thr629Met variant of case # 5 patient and 19 other pathogenic/likely pathogenic variants found in databases. (B) Human LeuRS molecular homology model, with the representation of domains and motifs. The zoom in shows variant analysis performed in three domains: light green for the catalytic (C,E), cyan for the editing (D) and orange for the leucine-specific (LS) (F) regions. For each domain mutations affect the electrostatic surface of the protein as well as the distance between neighboring residues. The p.Thr629Met variant lies in the LS domain between the hairpin of beta strand I–I, altering the folding of this loop and compromising the stability of the region (Zoom F). Detailed information regarding the genetic variants analyzed can be found in Table 3.